Secondary structure of the HIV reverse transcription initiation complex by NMR
- PMID: 21763492
- PMCID: PMC3710119
- DOI: 10.1016/j.jmb.2011.04.024
Secondary structure of the HIV reverse transcription initiation complex by NMR
Abstract
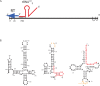
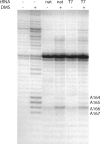
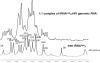
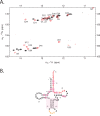
Initiation of reverse transcription of genomic RNA is a key early step in replication of the human immunodeficiency virus (HIV) upon infection of a host cell. Viral reverse transcriptase initiates from a specific RNA-RNA complex formed between a host transfer RNA (tRNA(Lys)(3)) and a region at the 5' end of genomic RNA; the 3' end of the tRNA acts as a primer for reverse transcription of genomic RNA. We report here the secondary structure of the HIV genomic RNA-human tRNA(Lys)(3) initiation complex using heteronuclear nuclear magnetic resonance methods. We show that both RNAs undergo large-scale conformational changes upon complex formation. Formation of the 18-bp primer helix with the 3' end of tRNA(Lys)(3) drives large conformational rearrangements of the tRNA at the 5' end while maintaining the anticodon loop for potential loop-loop interactions. HIV RNA forms an intramolecular helix adjacent to the intermolecular primer helix. This helix, which must be broken by reverse transcription, likely acts as a kinetic block to reverse transcription.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site.RNA. 2016 Nov;22(11):1689-1698. doi: 10.1261/rna.056804.116. Epub 2016 Sep 9. RNA. 2016. PMID: 27613581 Free PMC article.
-
Structural variability of the initiation complex of HIV-1 reverse transcription.J Biol Chem. 2004 Aug 20;279(34):35923-31. doi: 10.1074/jbc.M404473200. Epub 2004 Jun 11. J Biol Chem. 2004. PMID: 15194685
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
-
Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer).J Mol Biol. 1995 Mar 24;247(2):236-50. doi: 10.1006/jmbi.1994.0136. J Mol Biol. 1995. PMID: 7707372
-
Structural bases of the annealing of primer tRNA(3Lys) to the HIV-1 viral RNA.Curr HIV Res. 2005 Apr;3(2):147-56. doi: 10.2174/1570162053506919. Curr HIV Res. 2005. PMID: 15853720 Review.
Cited by
-
The Interaction between tRNA(Lys) 3 and the primer activation signal deciphered by NMR spectroscopy.PLoS One. 2013 Jun 6;8(6):e64700. doi: 10.1371/journal.pone.0064700. Print 2013. PLoS One. 2013. PMID: 23762248 Free PMC article.
-
Relating Structure and Dynamics in RNA Biology.Cold Spring Harb Perspect Biol. 2019 Jul 1;11(7):a032474. doi: 10.1101/cshperspect.a032474. Cold Spring Harb Perspect Biol. 2019. PMID: 31262948 Free PMC article. Review.
-
An unexpected RNA distal interaction mode found in an essential region of the hepatitis C virus genome.Nucleic Acids Res. 2018 May 4;46(8):4200-4212. doi: 10.1093/nar/gky074. Nucleic Acids Res. 2018. PMID: 29409065 Free PMC article.
-
Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.J Mol Biol. 2020 Jul 24;432(16):4499-4522. doi: 10.1016/j.jmb.2020.06.003. Epub 2020 Jun 6. J Mol Biol. 2020. PMID: 32512005 Free PMC article.
-
Dynamic Interplay of RNA and Protein in the Human Immunodeficiency Virus-1 Reverse Transcription Initiation Complex.J Mol Biol. 2018 Dec 7;430(24):5137-5150. doi: 10.1016/j.jmb.2018.08.029. Epub 2018 Sep 7. J Mol Biol. 2018. PMID: 30201267 Free PMC article.
References
-
- Barraud P, Gaudin C, Dardel F, Tisne C. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie. 2007 - PubMed
-
- Hu W-S, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. - PubMed
-
- Hsu M, Wainberg MA. Interactions between human immunodeficiency virus type 1 reverse transcriptase, tRNA primer, and nucleocapsid protein during reverse transcription. J Hum Virol. 2000;3:16–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

