HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation
- PMID: 21762802
- PMCID: PMC3139147
- DOI: 10.1016/j.jmb.2011.04.042
HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation
Abstract
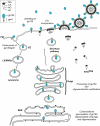
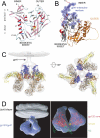
The HIV-1 envelope (Env) glycoproteins play an essential role in the virus replication cycle by mediating the fusion between viral and cellular membranes during the entry process. The Env glycoproteins are synthesized as a polyprotein precursor (gp160) that is cleaved by cellular proteases to the mature surface glycoprotein gp120 and the transmembrane glycoprotein gp41. During virus assembly, the gp120/gp41 complex is incorporated as heterotrimeric spikes into the lipid bilayer of nascent virions. These gp120/gp41 complexes then initiate the infection process by binding receptor and coreceptor on the surface of target cells. Much is currently known about the HIV-1 Env glycoprotein trafficking pathway and the structure of gp120 and the extracellular domain of gp41. However, the mechanism by which the Env glycoprotein complex is incorporated into virus particles remains incompletely understood. Genetic data support a major role for the cytoplasmic tail of gp41 and the matrix domain of Gag in Env glycoprotein incorporation. Still to be defined are the identities of host cell factors that may promote Env incorporation and the role of specific membrane microdomains in this process. Here, we review our current understanding of HIV-1 Env glycoprotein trafficking and incorporation into virions.
Published by Elsevier Ltd.
Figures


Similar articles
-
HIV-1 replication.Somat Cell Mol Genet. 2001 Nov;26(1-6):13-33. doi: 10.1023/a:1021070512287. Somat Cell Mol Genet. 2001. PMID: 12465460 Review.
-
Dual Pathways of Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Trafficking Modulate the Selective Exclusion of Uncleaved Oligomers from Virions.J Virol. 2021 Jan 13;95(3):e01369-20. doi: 10.1128/JVI.01369-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33148792 Free PMC article.
-
The Polar Region of the HIV-1 Envelope Protein Determines Viral Fusion and Infectivity by Stabilizing the gp120-gp41 Association.J Virol. 2019 Mar 21;93(7):e02128-18. doi: 10.1128/JVI.02128-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651369 Free PMC article.
-
Rab11-FIP1C Is Dispensable for HIV-1 Replication in Primary CD4+ T Cells, but Its Role Is Cell Type Dependent in Immortalized Human T-Cell Lines.J Virol. 2022 Dec 14;96(23):e0087622. doi: 10.1128/jvi.00876-22. Epub 2022 Nov 10. J Virol. 2022. PMID: 36354340 Free PMC article.
-
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation.Viruses. 2020 May 16;12(5):548. doi: 10.3390/v12050548. Viruses. 2020. PMID: 32429351 Free PMC article. Review.
Cited by
-
Optimized Infectivity of the Cell-Free Single-Cycle Human Immunodeficiency Viruses Type 1 (HIV-1) and Its Restriction by Host Cells.PLoS One. 2013 Jun 18;8(6):e67170. doi: 10.1371/journal.pone.0067170. Print 2013. PLoS One. 2013. PMID: 23825637 Free PMC article.
-
Detailed topology mapping reveals substantial exposure of the "cytoplasmic" C-terminal tail (CTT) sequences in HIV-1 Env proteins at the cell surface.PLoS One. 2013 May 27;8(5):e65220. doi: 10.1371/journal.pone.0065220. Print 2013. PLoS One. 2013. PMID: 23724133 Free PMC article.
-
C-terminal tail of human immunodeficiency virus gp41: functionally rich and structurally enigmatic.J Gen Virol. 2013 Jan;94(Pt 1):1-19. doi: 10.1099/vir.0.046508-0. Epub 2012 Oct 17. J Gen Virol. 2013. PMID: 23079381 Free PMC article. Review.
-
Interaction of TSG101 with the PTAP Motif in Distinct Locations of Gag Determines the Incorporation of HTLV-1 Env into the Retroviral Virion.Int J Mol Sci. 2023 Nov 20;24(22):16520. doi: 10.3390/ijms242216520. Int J Mol Sci. 2023. PMID: 38003710 Free PMC article.
-
Fangchinoline inhibits human immunodeficiency virus type 1 replication by interfering with gp160 proteolytic processing.PLoS One. 2012;7(6):e39225. doi: 10.1371/journal.pone.0039225. Epub 2012 Jun 13. PLoS One. 2012. PMID: 22720080 Free PMC article.
References
-
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
-
- Berger EA, Doms RW, Fenyö EM, Korber BT, Littman DR, Moore JP, Sattentau QJ, Schuitemaker H, Sodroski J, Weiss RA. A new classification for HIV-1. Nature. 1998;391:240. - PubMed
-
- Gorry PR, Ancuta P. Coreceptors and HIV-1 Pathogenesis. Curr HIV/AIDS Rep. 2011;8:45–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

