Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB
- PMID: 21760923
- PMCID: PMC3132758
- DOI: 10.1371/journal.pone.0021921
Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB
Abstract
Background: The phylum Crenarchaeota lacks the FtsZ cell division hallmark of bacteria and employs instead Cdv proteins. While CdvB and CdvC are homologues of the eukaryotic ESCRT-III and Vps4 proteins, implicated in membrane fission processes during multivesicular body biogenesis, cytokinesis and budding of some enveloped viruses, little is known about the structure and function of CdvA. Here, we report the biochemical and biophysical characterization of the three Cdv proteins from the hyperthermophilic archaeon Metallospherae sedula.
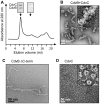
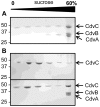
Methodology/principal findings: Using sucrose density gradient ultracentrifugation and negative staining electron microscopy, we evidenced for the first time that CdvA forms polymers in association with DNA, similar to known bacterial DNA partitioning proteins. We also observed that, in contrast to full-lengh CdvB that was purified as a monodisperse protein, the C-terminally deleted CdvB construct forms filamentous polymers, a phenomenon previously observed with eukaryotic ESCRT-III proteins. Based on size exclusion chromatography data combined with detection by multi-angle laser light scattering analysis, we demonstrated that CdvC assembles, in a nucleotide-independent way, as homopolymers resembling dodecamers and endowed with ATPase activity in vitro. The interactions between these putative cell division partners were further explored. Thus, besides confirming the previous observations that CdvB interacts with both CdvA and CdvC, our data demonstrate that CdvA/CdvB and CdvC/CdvB interactions are not mutually exclusive.
Conclusions/significance: Our data reinforce the concept that Cdv proteins are closely related to the eukaryotic ESCRT-III counterparts and suggest that the organization of the ESCRT-III machinery at the Crenarchaeal cell division septum is organized by CdvA an ancient cytoskeleton protein that might help to coordinate genome segregation.
Conflict of interest statement
Figures



Similar articles
-
Dividing the Archaeal Way: The Ancient Cdv Cell-Division Machinery.Front Microbiol. 2018 Mar 2;9:174. doi: 10.3389/fmicb.2018.00174. eCollection 2018. Front Microbiol. 2018. PMID: 29551994 Free PMC article. Review.
-
Deletion of cdvB paralogous genes of Sulfolobus acidocaldarius impairs cell division.Extremophiles. 2014 Mar;18(2):331-9. doi: 10.1007/s00792-013-0618-5. Epub 2014 Jan 8. Extremophiles. 2014. PMID: 24399085
-
The archaeal division protein CdvB1 assembles into polymers that are depolymerized by CdvC.FEBS Lett. 2022 Apr;596(7):958-969. doi: 10.1002/1873-3468.14324. Epub 2022 Mar 9. FEBS Lett. 2022. PMID: 35238034 Free PMC article.
-
The Structure, Function and Roles of the Archaeal ESCRT Apparatus.Subcell Biochem. 2017;84:357-377. doi: 10.1007/978-3-319-53047-5_12. Subcell Biochem. 2017. PMID: 28500532 Review.
-
The Nitrosopumilus maritimus CdvB, but not FtsZ, assembles into polymers.Archaea. 2013;2013:104147. doi: 10.1155/2013/104147. Epub 2013 Jun 2. Archaea. 2013. PMID: 23818813 Free PMC article.
Cited by
-
The ESCRT-III isoforms CHMP2A and CHMP2B display different effects on membranes upon polymerization.BMC Biol. 2021 Apr 8;19(1):66. doi: 10.1186/s12915-021-00983-9. BMC Biol. 2021. PMID: 33832485 Free PMC article.
-
Roles of ESCRT-III polymers in cell division across the tree of life.Curr Opin Cell Biol. 2023 Dec;85:102274. doi: 10.1016/j.ceb.2023.102274. Epub 2023 Nov 8. Curr Opin Cell Biol. 2023. PMID: 37944425 Free PMC article. Review.
-
Split decision: a thaumarchaeon encoding both FtsZ and Cdv cell division proteins chooses Cdv for cytokinesis.Mol Microbiol. 2011 Nov;82(3):535-8. doi: 10.1111/j.1365-2958.2011.07833.x. Epub 2011 Oct 10. Mol Microbiol. 2011. PMID: 21895799 Free PMC article.
-
Divided we stand: splitting synthetic cells for their proliferation.Syst Synth Biol. 2014 Sep;8(3):249-69. doi: 10.1007/s11693-014-9145-7. Epub 2014 May 27. Syst Synth Biol. 2014. PMID: 25136387 Free PMC article. Review.
-
The cell cycle of archaea.Nat Rev Microbiol. 2013 Sep;11(9):627-38. doi: 10.1038/nrmicro3077. Epub 2013 Jul 29. Nat Rev Microbiol. 2013. PMID: 23893102 Review.
References
-
- Garrett R, Klenk HP, editors. Archaea: evolution, physiology and molecular biology. Archaea ed. Blackwell Publishing; 2007.
-
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8:743–752. - PubMed
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

