The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli
- PMID: 21750663
- PMCID: PMC3130014
- DOI: 10.1371/journal.pbio.1001095
The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli
Abstract
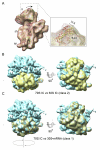
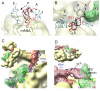
Formation of the 30S initiation complex (30S IC) is an important checkpoint in regulation of gene expression. The selection of mRNA, correct start codon, and the initiator fMet-tRNA(fMet) requires the presence of three initiation factors (IF1, IF2, IF3) of which IF3 and IF1 control the fidelity of the process, while IF2 recruits fMet-tRNA(fMet). Here we present a cryo-EM reconstruction of the complete 30S IC, containing mRNA, fMet-tRNA(fMet), IF1, IF2, and IF3. In the 30S IC, IF2 contacts IF1, the 30S subunit shoulder, and the CCA end of fMet-tRNA(fMet), which occupies a novel P/I position (P/I1). The N-terminal domain of IF3 contacts the tRNA, whereas the C-terminal domain is bound to the platform of the 30S subunit. Binding of initiation factors and fMet-tRNA(fMet) induces a rotation of the head relative to the body of the 30S subunit, which is likely to prevail through 50S subunit joining until GTP hydrolysis and dissociation of IF2 take place. The structure provides insights into the mechanism of mRNA selection during translation initiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
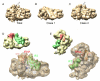


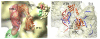

Similar articles
-
Structure of the 30S translation initiation complex.Nature. 2008 Sep 18;455(7211):416-20. doi: 10.1038/nature07192. Epub 2008 Aug 31. Nature. 2008. PMID: 18758445
-
Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria.Methods Enzymol. 2007;430:1-30. doi: 10.1016/S0076-6879(07)30001-3. Methods Enzymol. 2007. PMID: 17913632
-
Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.Cell. 2016 Sep 22;167(1):133-144.e13. doi: 10.1016/j.cell.2016.08.074. Cell. 2016. PMID: 27662086 Free PMC article.
-
A structural view of translation initiation in bacteria.Cell Mol Life Sci. 2009 Feb;66(3):423-36. doi: 10.1007/s00018-008-8416-4. Cell Mol Life Sci. 2009. PMID: 19011758 Free PMC article. Review.
-
Initiation of mRNA translation in bacteria: structural and dynamic aspects.Cell Mol Life Sci. 2015 Nov;72(22):4341-67. doi: 10.1007/s00018-015-2010-3. Epub 2015 Aug 11. Cell Mol Life Sci. 2015. PMID: 26259514 Free PMC article. Review.
Cited by
-
The initiation factor 3 (IF3) residues interacting with initiator tRNA elbow modulate the fidelity of translation initiation and growth fitness in Escherichia coli.Nucleic Acids Res. 2022 Nov 11;50(20):11712-11726. doi: 10.1093/nar/gkac1053. Nucleic Acids Res. 2022. PMID: 36399509 Free PMC article.
-
The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence.Proc Natl Acad Sci U S A. 2020 Jul 7;117(27):15565-15572. doi: 10.1073/pnas.1920013117. Epub 2020 Jun 23. Proc Natl Acad Sci U S A. 2020. PMID: 32576694 Free PMC article.
-
The emerging role of rectified thermal fluctuations in initiator aa-tRNA- and start codon selection during translation initiation.Biochimie. 2015 Jul;114:30-8. doi: 10.1016/j.biochi.2015.04.001. Epub 2015 Apr 14. Biochimie. 2015. PMID: 25882682 Free PMC article. Review.
-
Structure of Pseudomonas aeruginosa ribosomes from an aminoglycoside-resistant clinical isolate.Proc Natl Acad Sci U S A. 2019 Oct 29;116(44):22275-22281. doi: 10.1073/pnas.1909831116. Epub 2019 Oct 14. Proc Natl Acad Sci U S A. 2019. PMID: 31611393 Free PMC article.
-
Structural changes enable start codon recognition by the eukaryotic translation initiation complex.Cell. 2014 Oct 23;159(3):597-607. doi: 10.1016/j.cell.2014.10.001. Epub 2014 Oct 16. Cell. 2014. PMID: 25417110 Free PMC article.
References
-
- Gualerzi C. O, Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. - PubMed
-
- Studer S. M, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell. 2006;22:105–115. - PubMed
-
- Yusupova G. Z, Yusupov M. M, Cate J. H, Noller H. F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

