Down-regulation of EVI1 is associated with epigenetic alterations and good prognosis in patients with acute myeloid leukemia
- PMID: 21750091
- PMCID: PMC3186305
- DOI: 10.3324/haematol.2011.040535
Down-regulation of EVI1 is associated with epigenetic alterations and good prognosis in patients with acute myeloid leukemia
Abstract
Background: The EVI1 gene (3q26) codes for a zinc finger transcription factor with important roles in both mammalian development and leukemogenesis. Over-expression of EVI1 through either 3q26 rearrangements, MLL fusions, or other unknown mechanisms confers a poor prognosis in acute myeloid leukemia.
Design and methods: We analyzed the prevalence and prognostic impact of EVI1 over-expression in a series of 476 patients with acute myeloid leukemia, and investigated the epigenetic modifications of the EVI1 locus which could be involved in the transcriptional regulation of this gene.
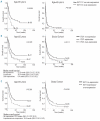
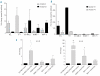
Results: Our data provide further evidence that EVI1 over-expression is a poor prognostic marker in acute myeloid leukemia patients less than 65 years old. Moreover, we found that patients with no basal expression of EVI1 had a better prognosis than patients with expression/over-expression (P=0.036). We also showed that cell lines with over-expression of EVI1 had no DNA methylation in the promoter region of the EVI1 locus, and had marks of active histone modifications: H3 and H4 acetylation, and trimethylation of histone H3 lysine 4. Conversely, cell lines with no expression of EVI1 have DNA hypermethylation and are marked by repressive trimethylation of histone H3 lysine 27 at the EVI1 promoter.
Conclusions: Our results identify EVI1 over-expression as a poor prognostic marker in a large, independent cohort of acute myeloid leukemia patients less than 65 years old, and show that the total absence of EVI1 expression has a prognostic impact on the outcome of such patients. Furthermore, we demonstrated for the first time that an aberrant epigenetic pattern involving DNA methylation, H3 and H4 acetylation, and trimethylation of histone H3 lysine 4 and histone H3 lysine 27 might play a role in the transcriptional regulation of EVI1 in acute myeloid leukemia. This study opens new avenues for a better understanding of the regulation of EVI1 expression at a transcriptional level.
Figures

Similar articles
-
Deregulated expression of EVI1 defines a poor prognostic subset of MLL-rearranged acute myeloid leukemias: a study of the German-Austrian Acute Myeloid Leukemia Study Group and the Dutch-Belgian-Swiss HOVON/SAKK Cooperative Group.J Clin Oncol. 2013 Jan 1;31(1):95-103. doi: 10.1200/JCO.2011.41.5505. Epub 2012 Sep 24. J Clin Oncol. 2013. PMID: 23008312
-
Expression and prognostic significance of different mRNA 5'-end variants of the oncogene EVI1 in 266 patients with de novo AML: EVI1 and MDS1/EVI1 overexpression both predict short remission duration.Genes Chromosomes Cancer. 2008 Apr;47(4):288-98. doi: 10.1002/gcc.20532. Genes Chromosomes Cancer. 2008. PMID: 18181178
-
EVI1 overexpression is a poor prognostic factor in pediatric patients with mixed lineage leukemia-AF9 rearranged acute myeloid leukemia.Haematologica. 2014 Nov;99(11):e225-7. doi: 10.3324/haematol.2014.107128. Epub 2014 Jul 11. Haematologica. 2014. PMID: 25015941 Free PMC article. No abstract available.
-
3q26/EVI1 rearrangements in myeloid hemopathies: a cytogenetic review.Future Oncol. 2015;11(11):1675-86. doi: 10.2217/fon.15.64. Future Oncol. 2015. PMID: 26043219 Review.
-
The role of EVI1 in myeloid malignancies.Blood Cells Mol Dis. 2014 Jun-Aug;53(1-2):67-76. doi: 10.1016/j.bcmd.2014.01.002. Epub 2014 Feb 1. Blood Cells Mol Dis. 2014. PMID: 24495476 Review.
Cited by
-
Orthogonal proteogenomic analysis identifies the druggable PA2G4-MYC axis in 3q26 AML.Nat Commun. 2024 Jun 4;15(1):4739. doi: 10.1038/s41467-024-48953-3. Nat Commun. 2024. PMID: 38834613 Free PMC article.
-
Noncoding RNAs in Acute Myeloid Leukemia: From Key Regulators to Clinical Players.Scientifica (Cairo). 2012;2012:925758. doi: 10.6064/2012/925758. Epub 2012 Sep 18. Scientifica (Cairo). 2012. PMID: 24278756 Free PMC article. Review.
-
High DNA methyltransferase DNMT3B levels: a poor prognostic marker in acute myeloid leukemia.PLoS One. 2012;7(12):e51527. doi: 10.1371/journal.pone.0051527. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251566 Free PMC article.
-
EVI‑1 acts as an oncogene and positively regulates calreticulin in breast cancer.Mol Med Rep. 2019 Mar;19(3):1645-1653. doi: 10.3892/mmr.2018.9796. Epub 2018 Dec 24. Mol Med Rep. 2019. Retraction in: Mol Med Rep. 2023 Dec;28(6):225. doi: 10.3892/mmr.2023.13112 PMID: 30592274 Free PMC article. Retracted.
-
Ecotropic viral integration site 1 regulates the progression of acute myeloid leukemia via MS4A3-mediated TGFβ/EMT signaling pathway.Oncol Lett. 2018 Aug;16(2):2701-2708. doi: 10.3892/ol.2018.8890. Epub 2018 Jun 4. Oncol Lett. 2018. PMID: 30013666 Free PMC article.
References
-
- Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene. 2006;368:1–11. - PubMed
-
- Wieser R. The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene. 2007;396(2):346–57. - PubMed
-
- Poppe B, Dastugue N, Vandesompele J, Cauwelier B, De Smet B, Yigit N, et al. EVI1 is consistently expressed as principal transcript in common and rare recurrent 3q26 rearrangements. Genes Chromosomes Cancer. 2006;45(4):349–56. - PubMed
-
- Lahortiga I, Vazquez I, Agirre X, Larrayoz MJ, Vizmanos JL, Gozzetti A, et al. Molecular heterogeneity in AML/MDS patients with 3q21q26 rearrangements. Genes Chromosomes Cancer. 2004;40(3):179–89. - PubMed

