Human linker histones: interplay between phosphorylation and O-β-GlcNAc to mediate chromatin structural modifications
- PMID: 21749719
- PMCID: PMC3149562
- DOI: 10.1186/1747-1028-6-15
Human linker histones: interplay between phosphorylation and O-β-GlcNAc to mediate chromatin structural modifications
Abstract
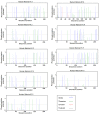
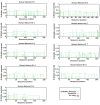
Eukaryotic chromatin is a combination of DNA and histone proteins. It is established fact that epigenetic mechanisms are associated with DNA and histones. Initial studies emphasize on core histones association with DNA, however later studies prove the importance of linker histone H1 epigenetic. There are many types of linker histone H1 found in mammals. These subtypes are cell specific and their amount in different types of cells varies as the cell functions. Many types of post-translational modifications which occur on different residues in each subtype of linker histone H1 induce conformational changes and allow the different subtypes of linker histone H1 to interact with chromatin at different stages during cell cycle which results in the regulation of transcription and gene expression. Proposed O-glycosylation of linker histone H1 promotes condensation of chromatin while phosphorylation of linker histone H1 is known to activate transcription and gene regulation by decondensation of chromatin. Interplay between phosphorylation and O-β-GlcNAc modification on Ser and Thr residues in each subtype of linker histone H1 in Homo sapiens during cell cycle may result in diverse functional regulation of proteins. This in silico study describes the potential phosphorylation, o-glycosylation and their possible interplay sites on conserved Ser/Thr residues in various subtypes of linker histone H1 in Homo sapiens.
Figures

Similar articles
-
Identification of novel post-translational modifications in linker histones from chicken erythrocytes.J Proteomics. 2015 Jan 15;113:162-77. doi: 10.1016/j.jprot.2014.10.004. Epub 2014 Oct 14. J Proteomics. 2015. PMID: 25452131
-
The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics.Biochim Biophys Acta. 2016 Mar;1859(3):486-95. doi: 10.1016/j.bbagrm.2015.09.003. Epub 2015 Sep 5. Biochim Biophys Acta. 2016. PMID: 26348411 Review.
-
Identification and Analysis of Six Phosphorylation Sites Within the Xenopus laevis Linker Histone H1.0 C-Terminal Domain Indicate Distinct Effects on Nucleosome Structure.Mol Cell Proteomics. 2022 Jul;21(7):100250. doi: 10.1016/j.mcpro.2022.100250. Epub 2022 May 23. Mol Cell Proteomics. 2022. PMID: 35618225 Free PMC article.
-
Dynamics and dispensability of variant-specific histone H1 Lys-26/Ser-27 and Thr-165 post-translational modifications.FEBS Lett. 2014 Jun 27;588(14):2353-62. doi: 10.1016/j.febslet.2014.05.035. Epub 2014 May 27. FEBS Lett. 2014. PMID: 24873882
-
Histone H1 and its isoforms: contribution to chromatin structure and function.Gene. 2009 Feb 15;431(1-2):1-12. doi: 10.1016/j.gene.2008.11.003. Epub 2008 Nov 14. Gene. 2009. PMID: 19059319 Review.
Cited by
-
O-GlcNAcylation: the sweet side of epigenetics.Epigenetics Chromatin. 2023 Dec 14;16(1):49. doi: 10.1186/s13072-023-00523-5. Epigenetics Chromatin. 2023. PMID: 38093337 Free PMC article. Review.
-
Modulation of epigenetic targets for anticancer therapy: clinicopathological relevance, structural data and drug discovery perspectives.Curr Pharm Des. 2013;19(4):578-613. doi: 10.2174/138161213804581918. Curr Pharm Des. 2013. PMID: 23016851 Free PMC article. Review.
-
Alternate Phosphorylation/O-GlcNAc Modification on Human Insulin IRSs: A Road towards Impaired Insulin Signaling in Alzheimer and Diabetes.Adv Bioinformatics. 2014;2014:324753. doi: 10.1155/2014/324753. Epub 2014 Dec 17. Adv Bioinformatics. 2014. PMID: 25580119 Free PMC article.
-
Nutrition, epigenetics, and metabolic syndrome.Antioxid Redox Signal. 2012 Jul 15;17(2):282-301. doi: 10.1089/ars.2011.4381. Epub 2012 Jan 13. Antioxid Redox Signal. 2012. PMID: 22044276 Free PMC article. Review.
-
Computer-aided Molecular Design of Compounds Targeting Histone Modifying Enzymes.Comput Struct Biotechnol J. 2015 May 7;13:358-65. doi: 10.1016/j.csbj.2015.04.007. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 26082827 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

