Influenza A viruses: new research developments
- PMID: 21747392
- PMCID: PMC10433403
- DOI: 10.1038/nrmicro2613
Influenza A viruses: new research developments
Abstract
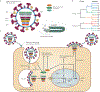
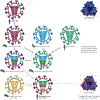
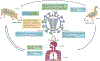
Influenza A viruses are zoonotic pathogens that continuously circulate and change in several animal hosts, including birds, pigs, horses and humans. The emergence of novel virus strains that are capable of causing human epidemics or pandemics is a serious possibility. Here, we discuss the value of surveillance and characterization of naturally occurring influenza viruses, and review the impact that new developments in the laboratory have had on our understanding of the host tropism and virulence of viruses. We also revise the lessons that have been learnt from the pandemic viruses of the past 100 years.
Figures

Similar articles
-
Virulence determinants of pandemic influenza viruses.J Clin Invest. 2011 Jan;121(1):6-13. doi: 10.1172/JCI44947. Epub 2011 Jan 4. J Clin Invest. 2011. PMID: 21206092 Free PMC article. Review.
-
Recent zoonoses caused by influenza A viruses.Rev Sci Tech. 2000 Apr;19(1):197-225. doi: 10.20506/rst.19.1.1220. Rev Sci Tech. 2000. PMID: 11189716 Review.
-
Influenza viruses: breaking all the rules.mBio. 2013 Jul 16;4(4):e00365-13. doi: 10.1128/mBio.00365-13. mBio. 2013. PMID: 23860766 Free PMC article.
-
[Interspecies transmission, adaptation to humans and pathogenicity of animal influenza viruses].Pathol Biol (Paris). 2010 Apr;58(2):e59-68. doi: 10.1016/j.patbio.2010.01.012. Epub 2010 Mar 19. Pathol Biol (Paris). 2010. PMID: 20303675 Review. French.
-
Predicting Zoonotic Risk of Influenza A Viruses from Host Tropism Protein Signature Using Random Forest.Int J Mol Sci. 2017 May 25;18(6):1135. doi: 10.3390/ijms18061135. Int J Mol Sci. 2017. PMID: 28587080 Free PMC article.
Cited by
-
CRISPR/Cas13a combined with hybridization chain reaction for visual detection of influenza A (H1N1) virus.Anal Bioanal Chem. 2022 Dec;414(29-30):8437-8445. doi: 10.1007/s00216-022-04380-1. Epub 2022 Oct 20. Anal Bioanal Chem. 2022. PMID: 36264297 Free PMC article.
-
An overview of the highly pathogenic H5N1 influenza virus.Virol Sin. 2013 Feb;28(1):3-15. doi: 10.1007/s12250-013-3294-9. Epub 2013 Jan 16. Virol Sin. 2013. PMID: 23325419 Free PMC article. Review.
-
Avian influenza A H5N1 virus: a continuous threat to humans.Emerg Microbes Infect. 2012 Sep;1(9):e25. doi: 10.1038/emi.2012.24. Epub 2012 Sep 19. Emerg Microbes Infect. 2012. PMID: 26038430 Free PMC article. Review.
-
Variable ligand- and receptor-binding hot spots in key strains of influenza neuraminidase.J Mol Genet Med. 2012;6:293-300. doi: 10.4172/1747-0862.1000052. Epub 2012 May 25. J Mol Genet Med. 2012. PMID: 22872804 Free PMC article.
-
Roles and functions of IAV proteins in host immune evasion.Front Immunol. 2023 Dec 13;14:1323560. doi: 10.3389/fimmu.2023.1323560. eCollection 2023. Front Immunol. 2023. PMID: 38152399 Free PMC article. Review.
References
-
- Molinari NA et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 (2007). - PubMed
-
- Johnson NP & Mueller J Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med 76, 105–115 (2002). - PubMed
-
- Palese P & Shaw ML in Fields Virology 5th edn (eds Knipe DM et al.) 1647–1689 (Lippincott Williams & Wilkins, Philadelphia, USA, 2007).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

