Probing for DNA damage with β-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro
- PMID: 21741328
- PMCID: PMC3212938
- DOI: 10.1016/j.dnarep.2011.04.020
Probing for DNA damage with β-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro
Abstract
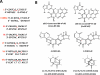
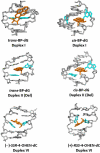
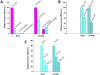
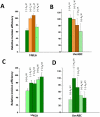
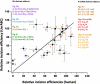
Nucleotide excision repair (NER) is an important prokaryotic and eukaryotic defense mechanism that removes a large variety of structurally distinct lesions in cellular DNA. While the proteins involved are completely different, the mode of action of these two repair systems is similar, involving a cut-and-patch mechanism in which an oligonucleotide sequence containing the lesion is excised. The prokaryotic and eukaryotic NER damage-recognition factors have common structural features of β-hairpin intrusion between the two DNA strands at the site of the lesion. In the present study, we explored the hypothesis that this common β-hairpin intrusion motif is mirrored in parallel NER incision efficiencies in the two systems. We have utilized human HeLa cell extracts and the prokaryotic UvrABC proteins to determine their relative NER incision efficiencies. We report here comparisons of relative NER efficiencies with a set of stereoisomeric DNA lesions derived from metabolites of benzo[a]pyrene and equine estrogens in different sequence contexts, utilizing 21 samples. We found a general qualitative trend toward similar relative NER incision efficiencies for ∼65% of these substrates; the other cases deviate mostly by ∼30% or less from a perfect correlation, although several more distant outliers are also evident. This resemblance is consistent with the hypothesis that lesion recognition through β-hairpin insertion, a common feature of the two systems, is facilitated by local thermodynamic destabilization induced by the lesions in both cases. In the case of the UvrABC system, varying the nature of the UvrC endonuclease, while maintaining the same UvrA/B proteins, can markedly affect the relative incision efficiencies. These observations suggest that, in addition to recognition involving the initial modified duplexes, downstream events involving UvrC can also play a role in distinguishing and processing different lesions in prokaryotic NER.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
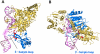

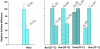

Similar articles
-
Reduced sulfhydryls maintain specific incision of BPDE-DNA adducts by recombinant thermoresistant Bacillus caldotenax UvrABC endonuclease.Protein Expr Purif. 2003 Sep;31(1):88-98. doi: 10.1016/s1046-5928(03)00137-2. Protein Expr Purif. 2003. PMID: 12963345
-
Robust incision of Benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system.Biochemistry. 2006 Jun 27;45(25):7834-43. doi: 10.1021/bi052515e. Biochemistry. 2006. PMID: 16784235 Free PMC article.
-
Exploring damage recognition models in prokaryotic nucleotide excision repair with a benzo[a]pyrene-derived lesion in UvrB.Biochemistry. 2009 Sep 29;48(38):8948-57. doi: 10.1021/bi9010072. Biochemistry. 2009. PMID: 19681599 Free PMC article.
-
The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch.Mutat Res. 2000 Aug 30;460(3-4):277-300. doi: 10.1016/s0921-8777(00)00032-x. Mutat Res. 2000. PMID: 10946234 Review.
-
Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery.Biopolymers. 2002 Nov 5;65(3):202-10. doi: 10.1002/bip.10239. Biopolymers. 2002. PMID: 12228925 Review.
Cited by
-
Recognition of Damaged DNA for Nucleotide Excision Repair: A Correlated Motion Mechanism with a Mismatched cis-syn Thymine Dimer Lesion.Biochemistry. 2015 Sep 1;54(34):5263-7. doi: 10.1021/acs.biochem.5b00840. Epub 2015 Aug 18. Biochemistry. 2015. PMID: 26270861 Free PMC article.
-
Prokaryotic nucleotide excision repair.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a012591. doi: 10.1101/cshperspect.a012591. Cold Spring Harb Perspect Biol. 2013. PMID: 23457260 Free PMC article. Review.
-
Adenine-DNA adducts derived from the highly tumorigenic Dibenzo[a,l]pyrene are resistant to nucleotide excision repair while guanine adducts are not.Chem Res Toxicol. 2013 May 20;26(5):783-93. doi: 10.1021/tx400080k. Epub 2013 Apr 24. Chem Res Toxicol. 2013. PMID: 23570232 Free PMC article.
-
The efficiencies of damage recognition and excision correlate with duplex destabilization induced by acetylaminofluorene adducts in human nucleotide excision repair.Chem Res Toxicol. 2012 Nov 19;25(11):2462-8. doi: 10.1021/tx3003033. Epub 2012 Oct 31. Chem Res Toxicol. 2012. PMID: 23088760 Free PMC article.
-
Structural and dynamic characterization of polymerase κ's minor groove lesion processing reveals how adduct topology impacts fidelity.Biochemistry. 2014 Sep 9;53(35):5683-91. doi: 10.1021/bi5007964. Epub 2014 Aug 22. Biochemistry. 2014. PMID: 25148552 Free PMC article.
References
-
- Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–52. - PubMed
-
- Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–76. - PubMed
-
- Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. - PubMed
-
- Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–98. - PubMed
-
- Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA099194-10/CA/NCI NIH HHS/United States
- CA28038/CA/NCI NIH HHS/United States
- R01 CA099194/CA/NCI NIH HHS/United States
- R01 CA112412-05S1/CA/NCI NIH HHS/United States
- R01 CA112412/CA/NCI NIH HHS/United States
- R01 CA028038/CA/NCI NIH HHS/United States
- R01 ES019566/ES/NIEHS NIH HHS/United States
- CA112412/CA/NCI NIH HHS/United States
- R01 CA075449-16/CA/NCI NIH HHS/United States
- C06RR-16572)/RR/NCRR NIH HHS/United States
- R01 CA028038-32/CA/NCI NIH HHS/United States
- CA75449/CA/NCI NIH HHS/United States
- R01 ES019566-02/ES/NIEHS NIH HHS/United States
- C06 RR016572/RR/NCRR NIH HHS/United States
- CA099194/CA/NCI NIH HHS/United States
- ES019566/ES/NIEHS NIH HHS/United States
- R01 CA075449/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

