Maps of protein structure space reveal a fundamental relationship between protein structure and function
- PMID: 21737750
- PMCID: PMC3145735
- DOI: 10.1073/pnas.1102727108
Maps of protein structure space reveal a fundamental relationship between protein structure and function
Abstract
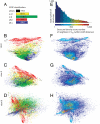
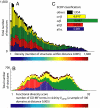
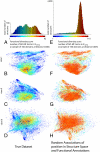
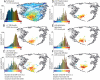
To study the protein structure-function relationship, we propose a method to efficiently create three-dimensional maps of structure space using a very large dataset of > 30,000 Structural Classification of Proteins (SCOP) domains. In our maps, each domain is represented by a point, and the distance between any two points approximates the structural distance between their corresponding domains. We use these maps to study the spatial distributions of properties of proteins, and in particular those of local vicinities in structure space such as structural density and functional diversity. These maps provide a unique broad view of protein space and thus reveal previously undescribed fundamental properties thereof. At the same time, the maps are consistent with previous knowledge (e.g., domains cluster by their SCOP class) and organize in a unified, coherent representation previous observation concerning specific protein folds. To investigate the function-structure relationship, we measure the functional diversity (using the Gene Ontology controlled vocabulary) in local structural vicinities. Our most striking finding is that functional diversity varies considerably across structure space: The space has a highly diverse region, and diversity abates when moving away from it. Interestingly, the domains in this region are mostly alpha/beta structures, which are known to be the most ancient proteins. We believe that our unique perspective of structure space will open previously undescribed ways of studying proteins, their evolution, and the relationship between their structure and function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Automatic classification of protein structures using low-dimensional structure space mappings.BMC Bioinformatics. 2014;15 Suppl 2(Suppl 2):S1. doi: 10.1186/1471-2105-15-S2-S1. Epub 2014 Jan 24. BMC Bioinformatics. 2014. PMID: 24564500 Free PMC article.
-
Cross-over between discrete and continuous protein structure space: insights into automatic classification and networks of protein structures.PLoS Comput Biol. 2009 Mar;5(3):e1000331. doi: 10.1371/journal.pcbi.1000331. Epub 2009 Mar 27. PLoS Comput Biol. 2009. PMID: 19325884 Free PMC article.
-
Global Mapping of Structures and Properties of Crystal Materials.J Chem Inf Model. 2023 Jun 26;63(12):3814-3826. doi: 10.1021/acs.jcim.3c00224. Epub 2023 Jun 13. J Chem Inf Model. 2023. PMID: 37310214 Review.
-
Navigating Among Known Structures in Protein Space.Methods Mol Biol. 2019;1851:233-249. doi: 10.1007/978-1-4939-8736-8_12. Methods Mol Biol. 2019. PMID: 30298400
-
Structural classification of proteins and structural genomics: new insights into protein folding and evolution.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Oct 1;66(Pt 10):1190-7. doi: 10.1107/S1744309110007177. Epub 2010 Jul 6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20944210 Free PMC article. Review.
Cited by
-
How to inherit statistically validated annotation within BAR+ protein clusters.BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2105-14-S3-S4. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514411 Free PMC article.
-
Identification and Analysis of Natural Building Blocks for Evolution-Guided Fragment-Based Protein Design.J Mol Biol. 2020 Jun 12;432(13):3898-3914. doi: 10.1016/j.jmb.2020.04.013. Epub 2020 Apr 21. J Mol Biol. 2020. PMID: 32330481 Free PMC article.
-
Functional and Structural Diversity of Acyl-coA Binding Proteins in Oil Crops.Front Genet. 2018 May 22;9:182. doi: 10.3389/fgene.2018.00182. eCollection 2018. Front Genet. 2018. PMID: 29872448 Free PMC article. Review.
-
Effective moment feature vectors for protein domain structures.PLoS One. 2013 Dec 31;8(12):e83788. doi: 10.1371/journal.pone.0083788. eCollection 2013. PLoS One. 2013. PMID: 24391828 Free PMC article.
-
Collision induced unfolding of isolated proteins in the gas phase: past, present, and future.Curr Opin Chem Biol. 2018 Feb;42:93-100. doi: 10.1016/j.cbpa.2017.11.010. Epub 2017 Dec 5. Curr Opin Chem Biol. 2018. PMID: 29207278 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

