Reductive evolution of bacterial genome in insect gut environment
- PMID: 21737395
- PMCID: PMC3157840
- DOI: 10.1093/gbe/evr064
Reductive evolution of bacterial genome in insect gut environment
Abstract
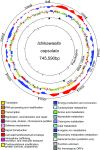
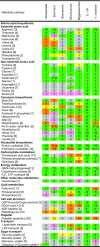
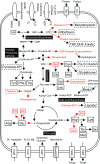
Obligate endocellular symbiotic bacteria of insects and other organisms generally exhibit drastic genome reduction. Recently, it was shown that symbiotic gut bacteria of some stinkbugs also have remarkably reduced genomes. Here, we report the complete genome sequence of such a gut bacterium Ishikawaella capsulata of the plataspid stinkbug Megacopta punctatissima. Gene repertoire and evolutionary patterns, including AT richness and elevated evolutionary rate, of the 745,590 bp genome were strikingly similar to those of obligate γ-proteobacterial endocellular insect symbionts like Buchnera in aphids and Wigglesworthia in tsetse flies. Ishikawaella was suggested to supply essential amino acids for the plant-sucking stinkbug as Buchnera does for the host aphid. Although Buchnera is phylogenetically closer to Wigglesworthia than to Ishikawaella, in terms of gene repertoire Buchnera was similar to Ishikawaella rather than to Wigglesworthia, providing a possible case of genome-level convergence of gene content. Meanwhile, several notable differences were identified between the genomes of Ishikawaella and Buchnera, including retention of TCA cycle genes and lack of flagellum-related genes in Ishikawaella, which may reflect their adaptation to distinct symbiotic habitats. Unexpectedly, Ishikawaella retained fewer genes related to cell wall synthesis and lipid metabolism than many endocellular insect symbionts. The plasmid of Ishikawaella encoded genes for arginine metabolism and oxalate detoxification, suggesting the possibility of additional Ishikawaella roles similar to those of human gut bacteria. Our data highlight strikingly similar evolutionary patterns that are shared between the extracellular and endocellular insect symbiont genomes.
Figures





Similar articles
-
Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria.PLoS Biol. 2006 Oct;4(10):e337. doi: 10.1371/journal.pbio.0040337. PLoS Biol. 2006. PMID: 17032065 Free PMC article.
-
Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs.BMC Biol. 2009 Jan 15;7:2. doi: 10.1186/1741-7007-7-2. BMC Biol. 2009. PMID: 19146674 Free PMC article.
-
Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches.BMC Biol. 2011 Dec 28;9:87. doi: 10.1186/1741-7007-9-87. BMC Biol. 2011. PMID: 22201529 Free PMC article.
-
Genome evolution in bacterial endosymbionts of insects.Nat Rev Genet. 2002 Nov;3(11):850-61. doi: 10.1038/nrg931. Nat Rev Genet. 2002. PMID: 12415315 Review.
-
The impact of microbial symbionts on host plant utilization by herbivorous insects.Mol Ecol. 2014 Mar;23(6):1473-1496. doi: 10.1111/mec.12421. Epub 2013 Aug 16. Mol Ecol. 2014. PMID: 23952067 Review.
Cited by
-
Demonstrating the role of symbionts in mediating detoxification in herbivores.Symbiosis. 2022;87(1):59-66. doi: 10.1007/s13199-022-00863-y. Epub 2022 Sep 13. Symbiosis. 2022. PMID: 36164313 Free PMC article.
-
Incipient genome erosion and metabolic streamlining for antibiotic production in a defensive symbiont.Proc Natl Acad Sci U S A. 2021 Apr 27;118(17):e2023047118. doi: 10.1073/pnas.2023047118. Proc Natl Acad Sci U S A. 2021. PMID: 33883280 Free PMC article.
-
How multi-partner endosymbioses function.Nat Rev Microbiol. 2016 Dec;14(12):731-743. doi: 10.1038/nrmicro.2016.151. Epub 2016 Oct 31. Nat Rev Microbiol. 2016. PMID: 27795568 Review.
-
Genome Sequence of "Candidatus Serratia symbiotica" Strain IS, a Facultative Bacterial Symbiont of the Pea Aphid Acyrthosiphon pisum.Microbiol Resour Announc. 2019 May 9;8(19):e00272-19. doi: 10.1128/MRA.00272-19. Microbiol Resour Announc. 2019. PMID: 31072900 Free PMC article.
-
The burrower bug Macroscytus japonensis (Hemiptera: Cydnidae) acquires obligate symbiotic bacteria from the environment.Zoological Lett. 2024 Aug 2;10(1):15. doi: 10.1186/s40851-024-00238-9. Zoological Lett. 2024. PMID: 39095847 Free PMC article.
References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Akman L, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. - PubMed
-
- Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. - PubMed
-
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

