The ribosome uses two active mechanisms to unwind messenger RNA during translation
- PMID: 21734708
- PMCID: PMC4170678
- DOI: 10.1038/nature10126
The ribosome uses two active mechanisms to unwind messenger RNA during translation
Abstract
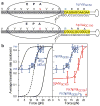
The ribosome translates the genetic information encoded in messenger RNA into protein. Folded structures in the coding region of an mRNA represent a kinetic barrier that lowers the peptide elongation rate, as the ribosome must disrupt structures it encounters in the mRNA at its entry site to allow translocation to the next codon. Such structures are exploited by the cell to create diverse strategies for translation regulation, such as programmed frameshifting, the modulation of protein expression levels, ribosome localization and co-translational protein folding. Although strand separation activity is inherent to the ribosome, requiring no exogenous helicases, its mechanism is still unknown. Here, using a single-molecule optical tweezers assay on mRNA hairpins, we find that the translation rate of identical codons at the decoding centre is greatly influenced by the GC content of folded structures at the mRNA entry site. Furthermore, force applied to the ends of the hairpin to favour its unfolding significantly speeds translation. Quantitative analysis of the force dependence of its helicase activity reveals that the ribosome, unlike previously studied helicases, uses two distinct active mechanisms to unwind mRNA structure: it destabilizes the helical junction at the mRNA entry site by biasing its thermal fluctuations towards the open state, increasing the probability of the ribosome translocating unhindered; and it mechanically pulls apart the mRNA single strands of the closed junction during the conformational changes that accompany ribosome translocation. The second of these mechanisms ensures a minimal basal rate of translation in the cell; specialized, mechanically stable structures are required to stall the ribosome temporarily. Our results establish a quantitative mechanical basis for understanding the mechanism of regulation of the elongation rate of translation by structured mRNAs.
©2011 Macmillan Publishers Limited. All rights reserved
Figures


Similar articles
-
Co-temporal Force and Fluorescence Measurements Reveal a Ribosomal Gear Shift Mechanism of Translation Regulation by Structured mRNAs.Mol Cell. 2019 Sep 5;75(5):1007-1019.e5. doi: 10.1016/j.molcel.2019.07.024. Epub 2019 Aug 27. Mol Cell. 2019. PMID: 31471187 Free PMC article.
-
Direct measurement of the mechanical work during translocation by the ribosome.Elife. 2014 Aug 11;3:e03406. doi: 10.7554/eLife.03406. Elife. 2014. PMID: 25114092 Free PMC article.
-
Single-Molecule Mechanical Folding and Unfolding of RNA Hairpins: Effects of Single A-U to A·C Pair Substitutions and Single Proton Binding and Implications for mRNA Structure-Induced -1 Ribosomal Frameshifting.J Am Chem Soc. 2018 Jul 5;140(26):8172-8184. doi: 10.1021/jacs.8b02970. Epub 2018 Jun 21. J Am Chem Soc. 2018. PMID: 29884019
-
Meanderings of the mRNA through the ribosome.Structure. 2001 Sep;9(9):751-8. doi: 10.1016/s0969-2126(01)00649-9. Structure. 2001. PMID: 11566123 Review.
-
How Messenger RNA and Nascent Chain Sequences Regulate Translation Elongation.Annu Rev Biochem. 2018 Jun 20;87:421-449. doi: 10.1146/annurev-biochem-060815-014818. Annu Rev Biochem. 2018. PMID: 29925264 Free PMC article. Review.
Cited by
-
Translation attenuation via 3' terminal codon usage in bovine csn1s2 is responsible for the difference in αs2- and β-casein profile in milk.RNA Biol. 2015;12(3):354-67. doi: 10.1080/15476286.2015.1017231. RNA Biol. 2015. PMID: 25826667 Free PMC article.
-
Human long noncoding RNAs are substantially less folded than messenger RNAs.Mol Biol Evol. 2015 Apr;32(4):970-7. doi: 10.1093/molbev/msu402. Epub 2014 Dec 23. Mol Biol Evol. 2015. PMID: 25540450 Free PMC article.
-
Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure.RNA. 2016 Jul;22(7):979-94. doi: 10.1261/rna.055251.115. Epub 2016 May 6. RNA. 2016. PMID: 27154968 Free PMC article.
-
Model of ribosome translation and mRNA unwinding.Eur Biophys J. 2013 May;42(5):347-54. doi: 10.1007/s00249-012-0879-4. Epub 2012 Dec 25. Eur Biophys J. 2013. PMID: 23266793
-
The ribosome in action: Tuning of translational efficiency and protein folding.Protein Sci. 2016 Aug;25(8):1390-406. doi: 10.1002/pro.2950. Epub 2016 Jun 8. Protein Sci. 2016. PMID: 27198711 Free PMC article. Review.
References
-
- Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. - PubMed
-
- Duan JB, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Human Molecular Genetics. 2003;12:205–216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

