Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells
- PMID: 21734045
- PMCID: PMC3165750
- DOI: 10.1128/JVI.00845-11
Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells
Abstract
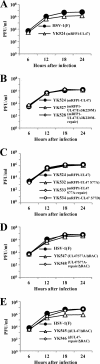
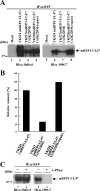
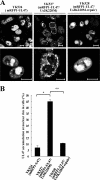
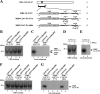
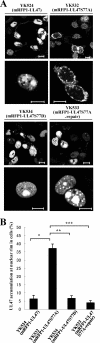
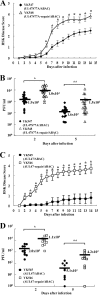
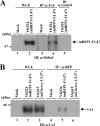
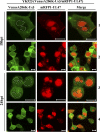
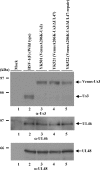
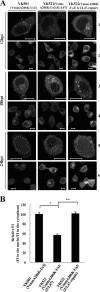
Us3 is a serine-threonine protein kinase encoded by herpes simplex virus 1 (HSV-1). We have identified UL47, a major virion protein, as a novel physiological substrate of Us3. In vitro kinase assays and systematic analysis of mutations at putative Us3 phosphorylation sites near the nuclear localization signal of UL47 showed that serine at residue 77 (Ser-77) was required for Us3 phosphorylation of UL47. Replacement of UL47 Ser-77 by alanine produced aberrant accumulation of UL47 at the nuclear rim and impaired the nuclear localization of UL47 in a significant fraction of infected cells. The same defect in UL47 localization was produced by an amino acid substitution in Us3 that inactivated its protein kinase activity. In contrast, a phosphomimetic mutation at UL47 Ser-77 restored wild-type nuclear localization. The UL47 S77A mutation also reduced viral replication in the mouse cornea and the development of herpes stromal keratitis in mice. In addition, UL47 formed a stable complex with Us3 in infected cells, and nuclear localization of Us3 was significantly impaired in the absence of UL47. These results suggested that Us3 phosphorylation of UL47 Ser-77 promoted the nuclear localization of UL47 in cell cultures and played a critical role in viral replication and pathogenesis in vivo. Furthermore, UL47 appeared to be required for efficient nuclear localization of Us3 in infected cells. Therefore, Us3 protein kinase and its substrate UL47 demonstrated a unique regulatory feature in that they reciprocally regulated their subcellular localization in infected cells.
Figures

Similar articles
-
Roles of Us8A and Its Phosphorylation Mediated by Us3 in Herpes Simplex Virus 1 Pathogenesis.J Virol. 2016 May 27;90(12):5622-5635. doi: 10.1128/JVI.00446-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27030266 Free PMC article.
-
Characterization of a Herpes Simplex Virus 1 (HSV-1) Chimera in Which the Us3 Protein Kinase Gene Is Replaced with the HSV-2 Us3 Gene.J Virol. 2015 Oct 21;90(1):457-73. doi: 10.1128/JVI.02376-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491159 Free PMC article.
-
Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery.J Virol. 2014 Mar;88(5):2775-85. doi: 10.1128/JVI.03300-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352467 Free PMC article.
-
Us3 Protein Kinase Encoded by HSV: The Precise Function and Mechanism on Viral Life Cycle.Adv Exp Med Biol. 2018;1045:45-62. doi: 10.1007/978-981-10-7230-7_3. Adv Exp Med Biol. 2018. PMID: 29896662 Review.
-
[Molecular mechanism by which Us3 protein kinase regulates the pathogenicity of herpes simplex virus type-1].Uirusu. 2016;66(1):83-90. doi: 10.2222/jsv.66.83. Uirusu. 2016. PMID: 28484184 Review. Japanese.
Cited by
-
DNA methyltransferase DNMT3A associates with viral proteins and impacts HSV-1 infection.Proteomics. 2015 Jun;15(12):1968-82. doi: 10.1002/pmic.201500035. Epub 2015 May 7. Proteomics. 2015. PMID: 25758154 Free PMC article.
-
Molecular characterization of duck enteritis virus UL41 protein.Virol J. 2018 Jan 15;15(1):12. doi: 10.1186/s12985-018-0928-4. Virol J. 2018. PMID: 29334975 Free PMC article.
-
Roles of Us8A and Its Phosphorylation Mediated by Us3 in Herpes Simplex Virus 1 Pathogenesis.J Virol. 2016 May 27;90(12):5622-5635. doi: 10.1128/JVI.00446-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27030266 Free PMC article.
-
Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence.J Virol. 2012 May;86(9):5264-77. doi: 10.1128/JVI.06913-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357273 Free PMC article.
-
US3 Kinase-Mediated Phosphorylation of Tegument Protein VP8 Plays a Critical Role in the Cellular Localization of VP8 and Its Effect on the Lipid Metabolism of Bovine Herpesvirus 1-Infected Cells.J Virol. 2019 Mar 5;93(6):e02151-18. doi: 10.1128/JVI.02151-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626671 Free PMC article.
References
-
- Asai R., Kato A., Kawaguchi Y. 2009. Epstein-Barr virus protein kinase BGLF4 interacts with viral transactivator BZLF1 and regulates its transactivation activity. J. Gen. Virol. 90:1575–1581 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

