The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites
- PMID: 21731489
- PMCID: PMC3121800
- DOI: 10.1371/journal.ppat.1002092
The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites
Abstract
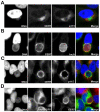
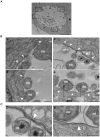
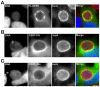
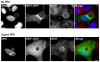
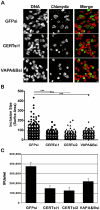
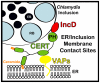
Bacterial pathogens that reside in membrane bound compartment manipulate the host cell machinery to establish and maintain their intracellular niche. The hijacking of inter-organelle vesicular trafficking through the targeting of small GTPases or SNARE proteins has been well established. Here, we show that intracellular pathogens also establish direct membrane contact sites with organelles and exploit non-vesicular transport machinery. We identified the ER-to-Golgi ceramide transfer protein CERT as a host cell factor specifically recruited to the inclusion, a membrane-bound compartment harboring the obligate intracellular pathogen Chlamydia trachomatis. We further showed that CERT recruitment to the inclusion correlated with the recruitment of VAPA/B-positive tubules in close proximity of the inclusion membrane, suggesting that ER-Inclusion membrane contact sites are formed upon C. trachomatis infection. Moreover, we identified the C. trachomatis effector protein IncD as a specific binding partner for CERT. Finally we showed that depletion of either CERT or the VAP proteins impaired bacterial development. We propose that the presence of IncD, CERT, VAPA/B, and potentially additional host and/or bacterial factors, at points of contact between the ER and the inclusion membrane provides a specialized metabolic and/or signaling microenvironment favorable to bacterial development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Rerouting of host lipids by bacteria: are you CERTain you need a vesicle?PLoS Pathog. 2011 Sep;7(9):e1002208. doi: 10.1371/journal.ppat.1002208. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909265 Free PMC article. No abstract available.
Similar articles
-
Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane.Infect Immun. 2014 May;82(5):2037-47. doi: 10.1128/IAI.01530-14. Epub 2014 Mar 4. Infect Immun. 2014. PMID: 24595143 Free PMC article.
-
Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development.PLoS Pathog. 2011 Sep;7(9):e1002198. doi: 10.1371/journal.ppat.1002198. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909260 Free PMC article.
-
STIM1 Is a Novel Component of ER-Chlamydia trachomatis Inclusion Membrane Contact Sites.PLoS One. 2015 Apr 27;10(4):e0125671. doi: 10.1371/journal.pone.0125671. eCollection 2015. PLoS One. 2015. PMID: 25915399 Free PMC article.
-
Chlamydiae interaction with the endoplasmic reticulum: contact, function and consequences.Cell Microbiol. 2015 Jul;17(7):959-66. doi: 10.1111/cmi.12455. Epub 2015 May 27. Cell Microbiol. 2015. PMID: 25930206 Free PMC article. Review.
-
Hijacking of Membrane Contact Sites by Intracellular Bacterial Pathogens.Adv Exp Med Biol. 2017;997:211-223. doi: 10.1007/978-981-10-4567-7_16. Adv Exp Med Biol. 2017. PMID: 28815533 Review.
Cited by
-
Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks.Prog Lipid Res. 2013 Jul;52(3):294-304. doi: 10.1016/j.plipres.2013.04.002. Epub 2013 Apr 19. Prog Lipid Res. 2013. PMID: 23608234 Free PMC article. Review.
-
Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19781-5. doi: 10.1073/pnas.1212831109. Epub 2012 Nov 5. Proc Natl Acad Sci U S A. 2012. PMID: 23129646 Free PMC article.
-
The chlamydial transcriptional regulator Euo is a key switch in cell form developmental progression but is not involved in the committed step to the formation of the infectious form.mSphere. 2024 Sep 25;9(9):e0043724. doi: 10.1128/msphere.00437-24. Epub 2024 Aug 14. mSphere. 2024. PMID: 39140730 Free PMC article.
-
Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates.J Bacteriol. 2012 Dec;194(23):6574-85. doi: 10.1128/JB.01428-12. Epub 2012 Oct 5. J Bacteriol. 2012. PMID: 23042990 Free PMC article.
-
Eukaryotic Clathrin Adapter Protein and Mediator of Cholesterol Homeostasis, PICALM, Affects Trafficking to the Chlamydial Inclusion.Mol Cell Biol. 2023 Feb 13;43(2):1-13. doi: 10.1080/10985549.2023.2171695. Online ahead of print. Mol Cell Biol. 2023. PMID: 36779337 Free PMC article.
References
-
- Dautry-Varsat A, Subtil A, Hackstadt T. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 2005;7:1714–1722. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

