microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs
- PMID: 21708909
- PMCID: PMC3153976
- DOI: 10.1261/rna.2775511
microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs
Abstract
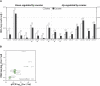
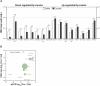
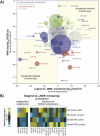
MicroRNAs (miRNAs) are small RNAs that modulate gene expression by binding target mRNAs. The hundreds of miRNAs expressed in the brain are critical for synaptic development and plasticity. Drugs of abuse cause lasting changes in the limbic regions of the brain that process reward, and addiction is viewed as a form of aberrant neuroplasticity. Using next-generation sequencing, we cataloged miRNA expression in the nucleus accumbens and at striatal synapses in control and chronically cocaine-treated mice. We identified cocaine-responsive miRNAs, synaptically enriched and depleted miRNA families, and confirmed cocaine-induced changes in protein expression for several predicted synaptic target genes. The miR-8 family, known for its roles in cancer, is highly enriched and cocaine regulated at striatal synapses, where its members may affect expression of cell adhesion molecules. Synaptically enriched cocaine-regulated miRNAs may contribute to long-lasting drug-induced plasticity through fine-tuning regulatory pathways that modulate the actin cytoskeleton, neurotransmitter metabolism, and peptide hormone processing.
Figures





Similar articles
-
microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity.Mol Cell Neurosci. 2009 Dec;42(4):350-62. doi: 10.1016/j.mcn.2009.08.009. Epub 2009 Aug 22. Mol Cell Neurosci. 2009. PMID: 19703567
-
Global Approaches to the Role of miRNAs in Drug-Induced Changes in Gene Expression.Front Genet. 2012 Jun 13;3:109. doi: 10.3389/fgene.2012.00109. eCollection 2012. Front Genet. 2012. PMID: 22707957 Free PMC article.
-
Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction.J Exp Med. 2010 Aug 30;207(9):1843-51. doi: 10.1084/jem.20100451. Epub 2010 Jul 19. J Exp Med. 2010. PMID: 20643829 Free PMC article.
-
[Regulation of microRNA Activity in Stress].Mol Biol (Mosk). 2017 Jul-Aug;51(4):561-572. doi: 10.7868/S0026898417030053. Mol Biol (Mosk). 2017. PMID: 28900074 Review. Russian.
-
Molecular, cellular, and structural mechanisms of cocaine addiction: a key role for microRNAs.Neuropsychopharmacology. 2013 Jan;38(1):198-211. doi: 10.1038/npp.2012.120. Epub 2012 Sep 12. Neuropsychopharmacology. 2013. PMID: 22968819 Free PMC article. Review.
Cited by
-
Microribonucleic acid dysregulations in children and adolescents with obsessive-compulsive disorder.Neuropsychiatr Dis Treat. 2015 Jul 14;11:1695-701. doi: 10.2147/NDT.S81884. eCollection 2015. Neuropsychiatr Dis Treat. 2015. PMID: 26203251 Free PMC article.
-
Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1.FASEB J. 2013 Sep;27(9):3720-9. doi: 10.1096/fj.13-232678. Epub 2013 Jun 10. FASEB J. 2013. PMID: 23752207 Free PMC article.
-
The Path to microRNA Therapeutics in Psychiatric and Neurodegenerative Disorders.Front Genet. 2012 May 17;3:82. doi: 10.3389/fgene.2012.00082. eCollection 2012. Front Genet. 2012. PMID: 22629284 Free PMC article.
-
mRNA changes in nucleus accumbens related to methamphetamine addiction in mice.Sci Rep. 2016 Nov 21;6:36993. doi: 10.1038/srep36993. Sci Rep. 2016. PMID: 27869204 Free PMC article.
-
Modulation by cocaine of dopamine receptors through miRNA-133b in zebrafish embryos.PLoS One. 2012;7(12):e52701. doi: 10.1371/journal.pone.0052701. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285158 Free PMC article.
References
-
- Betel D, Wilson M, Gabow A, Marks DS, Sander C 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153 - PMC - PubMed
-
- Bloomquist BT, Eipper BA, Mains RE 1991. Prohormone-converting enzymes: regulation and evaluation of function using antisense RNA. Mol Endocrinol 5: 2014–2024 - PubMed
-
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI 2007. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11). Biochemistry (Mosc) 72: 578–582 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
