Central and C-terminal domains of heterotrimeric G protein gamma subunits differentially influence the signaling necessary for primordial germ cell migration
- PMID: 21699975
- PMCID: PMC3303753
- DOI: 10.1016/j.cellsig.2011.05.015
Central and C-terminal domains of heterotrimeric G protein gamma subunits differentially influence the signaling necessary for primordial germ cell migration
Abstract
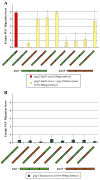
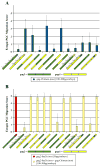
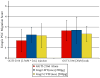
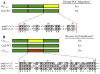
Heterotrimeric G protein signaling is involved in many pathways essential to development including those controlling cell migration, proliferation, differentiation and apoptosis. One key developmental event known to rely on proper heterotrimeric G protein signaling is primordial germ cell (PGC) migration. We previously developed an in vivo PGC migration assay that identified differences in the signaling capacity of G protein gamma subunits. In this study we developed Gγ subunit chimeras to determine the regions of Gγ isoforms that are responsible for these differences. The central section of the Gγ subunit was found to be necessary for the ability of a Gγ subunit to mediate signaling involved in PGC migration. Residues found in the carboxy-terminal segment of Gγ transducin (gngt1) were found to be responsible for the ability of this subunit to disrupt PGC migration. The type of prenylation did not affect the ability of a Gγ subunit to reverse prenylation-deficient-Gγ-induced PGC migration defects. However, a version of gng2, engineered to be farnesylated instead of geranylgeranylated, still lacks the ability to reverse PGC migration defects known to result from treatment of zebrafish with geranylgeranyl transferase inhibitors (GGTI), supporting the notion that Gγ subunits are one of several protein targets that need to be geranylgeranylated to orchestrate the proper long-range migration of PGCs.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Prenylation-deficient G protein gamma subunits disrupt GPCR signaling in the zebrafish.Cell Signal. 2010 Feb;22(2):221-33. doi: 10.1016/j.cellsig.2009.09.017. Epub 2009 Sep 26. Cell Signal. 2010. PMID: 19786091 Free PMC article.
-
Gγ identity dictates efficacy of Gβγ signaling and macrophage migration.J Biol Chem. 2018 Feb 23;293(8):2974-2989. doi: 10.1074/jbc.RA117.000872. Epub 2018 Jan 9. J Biol Chem. 2018. PMID: 29317505 Free PMC article.
-
Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells.Dev Biol. 2006 Apr 15;292(2):393-406. doi: 10.1016/j.ydbio.2006.01.014. Epub 2006 Mar 2. Dev Biol. 2006. PMID: 16513105
-
Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding.Biochem Pharmacol. 2001 Jun 1;61(11):1329-37. doi: 10.1016/s0006-2952(01)00633-5. Biochem Pharmacol. 2001. PMID: 11331068 Review.
-
Mechanisms guiding primordial germ cell migration: strategies from different organisms.Nat Rev Mol Cell Biol. 2010 Jan;11(1):37-49. doi: 10.1038/nrm2815. Nat Rev Mol Cell Biol. 2010. PMID: 20027186 Free PMC article. Review.
Cited by
-
Endothelin-1 Pathway Polymorphisms and Outcomes in Pulmonary Arterial Hypertension.Am J Respir Crit Care Med. 2015 Dec 1;192(11):1345-54. doi: 10.1164/rccm.201501-0196OC. Am J Respir Crit Care Med. 2015. PMID: 26252367 Free PMC article.
-
Identified GNGT1 and NMU as Combined Diagnosis Biomarker of Non-Small-Cell Lung Cancer Utilizing Bioinformatics and Logistic Regression.Dis Markers. 2021 Jan 6;2021:6696198. doi: 10.1155/2021/6696198. eCollection 2021. Dis Markers. 2021. PMID: 33505535 Free PMC article.
-
Identification of differentially expressed genes during development of the zebrafish pineal complex using RNA sequencing.Dev Biol. 2014 Nov 1;395(1):144-53. doi: 10.1016/j.ydbio.2014.08.015. Epub 2014 Aug 28. Dev Biol. 2014. PMID: 25173875 Free PMC article.
-
Maternal Hypermethylated Genes Contribute to Intrauterine Growth Retardation of Piglets in Rongchang Pigs.Int J Mol Sci. 2024 Jun 12;25(12):6462. doi: 10.3390/ijms25126462. Int J Mol Sci. 2024. PMID: 38928167 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

