Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
- PMID: 21685874
- PMCID: PMC3160252
- DOI: 10.1038/emboj.2011.193
Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Abstract
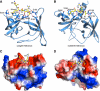
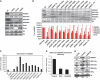
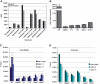
The SAGA (Spt-Ada-Gcn5 acetyltransferase) complex is an important chromatin modifying complex that can both acetylate and deubiquitinate histones. Sgf29 is a novel component of the SAGA complex. Here, we report the crystal structures of the tandem Tudor domains of Saccharomyces cerevisiae and human Sgf29 and their complexes with H3K4me2 and H3K4me3 peptides, respectively, and show that Sgf29 selectively binds H3K4me2/3 marks. Our crystal structures reveal that Sgf29 harbours unique tandem Tudor domains in its C-terminus. The tandem Tudor domains in Sgf29 tightly pack against each other face-to-face with each Tudor domain harbouring a negatively charged pocket accommodating the first residue alanine and methylated K4 residue of histone H3, respectively. The H3A1 and K4me3 binding pockets and the limited binding cleft length between these two binding pockets are the structural determinants in conferring the ability of Sgf29 to selectively recognize H3K4me2/3. Our in vitro and in vivo functional assays show that Sgf29 recognizes methylated H3K4 to recruit the SAGA complex to its targets sites and mediates histone H3 acetylation, underscoring the importance of Sgf29 in gene regulation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

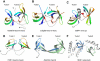

Similar articles
-
Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation.Nature. 2005 Jan 27;433(7024):434-8. doi: 10.1038/nature03242. Epub 2005 Jan 12. Nature. 2005. PMID: 15647753
-
Nucleosome competition reveals processive acetylation by the SAGA HAT module.Proc Natl Acad Sci U S A. 2015 Oct 6;112(40):E5461-70. doi: 10.1073/pnas.1508449112. Epub 2015 Sep 23. Proc Natl Acad Sci U S A. 2015. PMID: 26401015 Free PMC article.
-
The N-terminus and Tudor domains of Sgf29 are important for its heterochromatin boundary formation function.J Biochem. 2014 Mar;155(3):159-71. doi: 10.1093/jb/mvt108. Epub 2013 Dec 3. J Biochem. 2014. PMID: 24307402
-
The SAGA continues: The rise of cis- and trans-histone crosstalk pathways.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194600. doi: 10.1016/j.bbagrm.2020.194600. Epub 2020 Jul 6. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32645359 Free PMC article. Review.
-
The Ada2/Ada3/Gcn5/Sgf29 histone acetyltransferase module.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194629. doi: 10.1016/j.bbagrm.2020.194629. Epub 2020 Sep 2. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32890768 Free PMC article. Review.
Cited by
-
Therapeutic targeting Tudor domains in leukemia via CRISPR-Scan Assisted Drug Discovery.Sci Adv. 2024 Feb 23;10(8):eadk3127. doi: 10.1126/sciadv.adk3127. Epub 2024 Feb 23. Sci Adv. 2024. PMID: 38394203 Free PMC article.
-
Loss of the Ash2l subunit of histone H3K4 methyltransferase complexes reduces chromatin accessibility at promoters.Sci Rep. 2022 Dec 13;12(1):21506. doi: 10.1038/s41598-022-25881-0. Sci Rep. 2022. PMID: 36513698 Free PMC article.
-
MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.Acta Crystallogr F Struct Biol Commun. 2018 Sep 1;74(Pt 9):549-557. doi: 10.1107/S2053230X18009901. Epub 2018 Aug 29. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30198887 Free PMC article.
-
Conserved and plant-specific histone acetyltransferase complexes cooperate to regulate gene transcription and plant development.Nat Plants. 2023 Mar;9(3):442-459. doi: 10.1038/s41477-023-01359-3. Epub 2023 Mar 6. Nat Plants. 2023. PMID: 36879016
-
A synthetic non-histone substrate to study substrate targeting by the Gcn5 HAT and sirtuin HDACs.J Biol Chem. 2019 Apr 19;294(16):6227-6239. doi: 10.1074/jbc.RA118.006051. Epub 2019 Feb 25. J Biol Chem. 2019. PMID: 30804216 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

