PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1
- PMID: 21677187
- PMCID: PMC3610090
- DOI: 10.1523/JNEUROSCI.5410-10.2011
PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1
Abstract
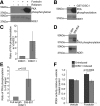
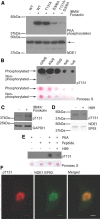
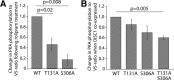
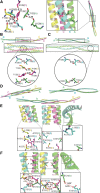
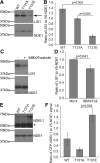
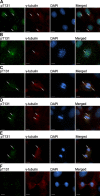
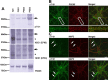
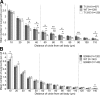
Nuclear distribution factor E-homolog 1 (NDE1), Lissencephaly 1 (LIS1), and NDE-like 1 (NDEL1) together participate in essential neurodevelopmental processes, including neuronal precursor proliferation and differentiation, neuronal migration, and neurite outgrowth. NDE1/LIS1/NDEL1 interacts with Disrupted in Schizophrenia 1 (DISC1) and the cAMP-hydrolyzing enzyme phosphodiesterase 4 (PDE4). DISC1, PDE4, NDE1, and NDEL1 have each been implicated as genetic risk factors for major mental illness. Here, we demonstrate that DISC1 and PDE4 modulate NDE1 phosphorylation by cAMP-dependent protein kinase A (PKA) and identify a novel PKA substrate site on NDE1 at threonine-131 (T131). Homology modeling predicts that phosphorylation at T131 modulates NDE1-LIS1 and NDE1-NDEL1 interactions, which we confirm experimentally. DISC1-PDE4 interaction thus modulates organization of the NDE1/NDEL1/LIS1 complex. T131-phosphorylated NDE1 is present at the postsynaptic density, in proximal axons, within the nucleus, and at the centrosome where it becomes substantially enriched during mitosis. Mutation of the NDE1 T131 site to mimic PKA phosphorylation inhibits neurite outgrowth. Thus PKA-dependent phosphorylation of the NDE1/LIS1/NDEL1 complex is DISC1-PDE4 modulated and likely to regulate its neural functions.
Figures
Similar articles
-
DISC1, PDE4B, and NDE1 at the centrosome and synapse.Biochem Biophys Res Commun. 2008 Dec 26;377(4):1091-6. doi: 10.1016/j.bbrc.2008.10.120. Epub 2008 Nov 5. Biochem Biophys Res Commun. 2008. PMID: 18983980
-
NDE1 and NDEL1: multimerisation, alternate splicing and DISC1 interaction.Neurosci Lett. 2009 Jan 16;449(3):228-33. doi: 10.1016/j.neulet.2008.10.095. Epub 2008 Nov 6. Neurosci Lett. 2009. PMID: 19000741 Free PMC article.
-
[When we have learned about the brain development from a disease-oriented study: DBZ regulates cortical cell positioning and neurite extension by sustaining the anterograde transport of Lis1/DISC1 through control of Ndel1 phosphorylation].Nihon Shinkei Seishin Yakurigaku Zasshi. 2016 Apr;36(2):43-50. Nihon Shinkei Seishin Yakurigaku Zasshi. 2016. PMID: 27333658 Review. Japanese.
-
DBZ regulates cortical cell positioning and neurite development by sustaining the anterograde transport of Lis1 and DISC1 through control of Ndel1 dual-phosphorylation.J Neurosci. 2015 Feb 18;35(7):2942-58. doi: 10.1523/JNEUROSCI.5029-13.2015. J Neurosci. 2015. PMID: 25698733 Free PMC article.
-
Protein-Protein and Peptide-Protein Interactions of NudE-Like 1 (Ndel1): A Protein Involved in Schizophrenia.Curr Protein Pept Sci. 2015;16(8):754-67. doi: 10.2174/1389203716666150505225251. Curr Protein Pept Sci. 2015. PMID: 25961396 Review.
Cited by
-
Interneuron Heterotopia in the Lis1 Mutant Mouse Cortex Underlies a Structural and Functional Schizophrenia-Like Phenotype.Front Cell Dev Biol. 2021 Jul 13;9:693919. doi: 10.3389/fcell.2021.693919. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34327202 Free PMC article.
-
Lead Neurotoxicity on Human Neuroblastoma Cell Line SH-SY5Y is Mediated via Transcription Factor EGR1/Zif268 Induced Disrupted in Scherophernia-1 Activation.Neurochem Res. 2018 Jul;43(7):1308-1316. doi: 10.1007/s11064-018-2539-2. Epub 2018 Jun 4. Neurochem Res. 2018. PMID: 29868983
-
Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway.J Cell Biol. 2016 Feb 15;212(4):409-23. doi: 10.1083/jcb.201507046. J Cell Biol. 2016. PMID: 26880200 Free PMC article.
-
DISC1 genetics, biology and psychiatric illness.Front Biol (Beijing). 2013 Feb 1;8(1):1-31. doi: 10.1007/s11515-012-1254-7. Front Biol (Beijing). 2013. PMID: 23550053 Free PMC article.
-
Dominant-Negative Attenuation of cAMP-Selective Phosphodiesterase PDE4D Action Affects Learning and Behavior.Int J Mol Sci. 2020 Aug 9;21(16):5704. doi: 10.3390/ijms21165704. Int J Mol Sci. 2020. PMID: 32784895 Free PMC article.
References
-
- Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–1096. - PubMed
-
- Brandon NJ, Handford EJ, Schurov I, Rain J-C, Pelling M, Duran-Jimeriz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. - PubMed
-
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, Malhotra AK. Elucidating the relationship between DISC1, NDEL1, and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
