Amino acid precursor supply in the biosynthesis of the RNA polymerase inhibitor streptolydigin by Streptomyces lydicus
- PMID: 21665968
- PMCID: PMC3147693
- DOI: 10.1128/JB.05062-11
Amino acid precursor supply in the biosynthesis of the RNA polymerase inhibitor streptolydigin by Streptomyces lydicus
Abstract
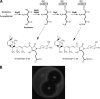
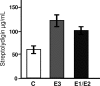
Biosynthesis of the hybrid polyketide-nonribosomal peptide antibiotic streptolydigin, 3-methylaspartate, is utilized as precursor of the tetramic acid moiety. The three genes from the Streptomyces lydicus streptolydigin gene cluster slgE1-slgE2-slgE3 are involved in 3-methylaspartate supply. SlgE3, a ferredoxin-dependent glutamate synthase, is responsible for the biosynthesis of glutamate from glutamine and 2-oxoglutarate. In addition to slgE3, housekeeping NADPH- and ferredoxin-dependent glutamate synthase genes have been identified in S. lydicus. The expression of slgE3 is increased up to 9-fold at the onset of streptolydigin biosynthesis and later decreases to ∼2-fold over the basal level. In contrast, the expression of housekeeping glutamate synthases decreases when streptolydigin begins to be synthesized. SlgE1 and SlgE2 are the two subunits of a glutamate mutase that would convert glutamate into 3-methylaspartate. Deletion of slgE1-slgE2 led to the production of two compounds containing a lateral side chain derived from glutamate instead of 3-methylaspartate. Expression of this glutamate mutase also reaches a peak increase of up to 5.5-fold coinciding with the onset of antibiotic production. Overexpression of either slgE3 or slgE1-slgE2 in S. lydicus led to an increase in the yield of streptolydigin.
Figures

Similar articles
-
Insights into the roles of exogenous glutamate and proline in improving streptolydigin production of Streptomyces lydicus with metabolomic analysis.J Ind Microbiol Biotechnol. 2013 Nov;40(11):1303-14. doi: 10.1007/s10295-013-1326-y. Epub 2013 Aug 29. J Ind Microbiol Biotechnol. 2013. PMID: 23990132
-
Three pathway-specific regulators control streptolydigin biosynthesis in Streptomyces lydicus.Microbiology (Reading). 2012 Oct;158(Pt 10):2504-2514. doi: 10.1099/mic.0.061325-0. Epub 2012 Jul 19. Microbiology (Reading). 2012. PMID: 22820841
-
Novel compounds produced by Streptomyces lydicus NRRL 2433 engineered mutants altered in the biosynthesis of streptolydigin.J Antibiot (Tokyo). 2012 Jul;65(7):341-8. doi: 10.1038/ja.2012.37. Epub 2012 May 9. J Antibiot (Tokyo). 2012. PMID: 22569159
-
Biosynthesis of the RNA polymerase inhibitor streptolydigin in Streptomyces lydicus: tailoring modification of 3-methyl-aspartate.J Bacteriol. 2011 May;193(10):2647-51. doi: 10.1128/JB.00108-11. Epub 2011 Mar 11. J Bacteriol. 2011. PMID: 21398531 Free PMC article.
-
fabC of Streptomyces lydicus involvement in the biosynthesis of streptolydigin.Appl Microbiol Biotechnol. 2009 May;83(2):305-13. doi: 10.1007/s00253-009-1872-4. Epub 2009 Feb 13. Appl Microbiol Biotechnol. 2009. PMID: 19214500
Cited by
-
Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes.J Biol Chem. 2013 Jan 18;288(3):1991-2003. doi: 10.1074/jbc.M112.420182. Epub 2012 Nov 28. J Biol Chem. 2013. PMID: 23192349 Free PMC article.
-
Manipulating natural product biosynthetic pathways via DNA assembler.Curr Protoc Chem Biol. 2014 Jun 3;6(2):65-100. doi: 10.1002/9780470559277.ch130191. Curr Protoc Chem Biol. 2014. PMID: 24903884 Free PMC article.
-
Enhancement of precursor amino acid supplies for improving bacitracin production by activation of branched chain amino acid transporter BrnQ and deletion of its regulator gene lrp in Bacillus licheniformis.Synth Syst Biotechnol. 2018 Nov 2;3(4):236-243. doi: 10.1016/j.synbio.2018.10.009. eCollection 2018 Dec. Synth Syst Biotechnol. 2018. PMID: 30417137 Free PMC article.
-
Insights into the roles of exogenous glutamate and proline in improving streptolydigin production of Streptomyces lydicus with metabolomic analysis.J Ind Microbiol Biotechnol. 2013 Nov;40(11):1303-14. doi: 10.1007/s10295-013-1326-y. Epub 2013 Aug 29. J Ind Microbiol Biotechnol. 2013. PMID: 23990132
-
Complete genome sequencing and antibiotics biosynthesis pathways analysis of Streptomyces lydicus 103.Sci Rep. 2017 Mar 20;7:44786. doi: 10.1038/srep44786. Sci Rep. 2017. PMID: 28317865 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

