Autophagy and p62 in cardiac proteinopathy
- PMID: 21659648
- PMCID: PMC3142307
- DOI: 10.1161/CIRCRESAHA.111.244707
Autophagy and p62 in cardiac proteinopathy
Abstract
Rationale: Recent studies suggest an important role of autophagy in protection against αB-crystallin-based (CryAB(R120G)) desmin-related cardiomyopathies (DRC), but this has not been demonstrated in a different model of cardiac proteinopathy. Mechanisms underlying the response of cardiomyocytes to proteotoxic stress remain incompletely understood.
Objective: Our first objective was to determine whether and how the autophagic activity is changed in a mouse model of desminopathy. We also investigated the role of p62 in the protein quality control of cardiomyocytes.
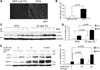
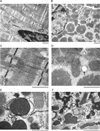
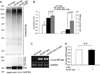
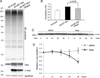
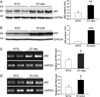
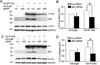
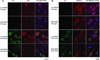
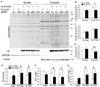
Methods and results: Using an autophagosome reporter and determining changes in LC3-II protein levels in response to lysosomal inhibition, we found significantly increased autophagic flux in mouse hearts with transgenic overexpression of a DRC-linked mutant desmin. Similarly, autophagic flux was increased in cultured neonatal rat ventricular myocytes (NRVMs) expressing a mutant desmin. Suppression of autophagy by 3-methyladenine increased, whereas enhancement of autophagy by rapamycin reduced the ability of a comparable level of mutant desmin overexpression to accumulate ubiquitinated proteins in NRVMs. Furthermore, p62 mRNA and protein expression was significantly up-regulated in cardiomyocytes by transgenic overexpression of the mutant desmin or CryAB(R120G) both in intact mice and in vitro. The p62 depletion impaired aggresome and autophagosome formation, exacerbated cell injury, and decreased cell viability in cultured NRVMs expressing the misfolded proteins.
Conclusions: Autophagic flux is increased in desminopathic hearts, and as previously suggested in CryAB(R120G)-based DRC, this increased autophagic flux serves as an adaptive response to overexpression of misfolded proteins. The p62 is up-regulated in mouse proteinopathic hearts. The p62 promotes aggresome formation and autophagy activation and protects cardiomyocytes against proteotoxic stress.
Figures
Similar articles
-
Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes.Circ Res. 2011 Jul 8;109(2):151-60. doi: 10.1161/CIRCRESAHA.110.237339. Epub 2011 May 26. Circ Res. 2011. PMID: 21617129 Free PMC article.
-
Doxycycline attenuates protein aggregation in cardiomyocytes and improves survival of a mouse model of cardiac proteinopathy.J Am Coll Cardiol. 2010 Oct 19;56(17):1418-26. doi: 10.1016/j.jacc.2010.01.075. J Am Coll Cardiol. 2010. PMID: 20947000 Free PMC article.
-
TFEB activation protects against cardiac proteotoxicity via increasing autophagic flux.J Mol Cell Cardiol. 2017 Dec;113:51-62. doi: 10.1016/j.yjmcc.2017.10.003. Epub 2017 Oct 7. J Mol Cell Cardiol. 2017. PMID: 28993153 Free PMC article.
-
Desmin filaments and cardiac disease: establishing causality.J Card Fail. 2002 Dec;8(6 Suppl):S287-92. doi: 10.1054/jcaf.2002.129279. J Card Fail. 2002. PMID: 12555134 Review.
-
p62 Stages an interplay between the ubiquitin-proteasome system and autophagy in the heart of defense against proteotoxic stress.Trends Cardiovasc Med. 2011 Nov;21(8):224-8. doi: 10.1016/j.tcm.2012.05.015. Trends Cardiovasc Med. 2011. PMID: 22902070 Free PMC article. Review.
Cited by
-
Involvement of autophagy in cardiac remodeling in transgenic mice with cardiac specific over-expression of human programmed cell death 5.PLoS One. 2012;7(1):e30097. doi: 10.1371/journal.pone.0030097. Epub 2012 Jan 11. PLoS One. 2012. PMID: 22253891 Free PMC article.
-
Cardiac fibrosis in mouse expressing DsRed tetramers involves chronic autophagy and proteasome degradation insufficiency.Oncotarget. 2016 Aug 23;7(34):54274-54289. doi: 10.18632/oncotarget.11026. Oncotarget. 2016. PMID: 27494843 Free PMC article.
-
WX20120108, a novel IAP antagonist, induces tumor cell autophagy via activating ROS-FOXO pathway.Acta Pharmacol Sin. 2019 Nov;40(11):1466-1479. doi: 10.1038/s41401-019-0253-5. Epub 2019 Jul 17. Acta Pharmacol Sin. 2019. PMID: 31316176 Free PMC article.
-
Systemic inhibition of neddylation by 3-day MLN4924 treatment regime does not impair autophagic flux in mouse hearts and brains.Am J Cardiovasc Dis. 2017 Dec 20;7(6):134-150. eCollection 2017. Am J Cardiovasc Dis. 2017. PMID: 29348974 Free PMC article.
-
Fibin regulates cardiomyocyte hypertrophy and causes protein-aggregate-associated cardiomyopathy in vivo.Front Mol Biosci. 2023 Jun 5;10:1169658. doi: 10.3389/fmolb.2023.1169658. eCollection 2023. Front Mol Biosci. 2023. PMID: 37342207 Free PMC article.
References
-
- Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. - PubMed
-
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. - PubMed
-
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL085629-02/HL/NHLBI NIH HHS/United States
- R01 HL072166-06A1S1/HL/NHLBI NIH HHS/United States
- R01 HL072166-05/HL/NHLBI NIH HHS/United States
- R01 HL072166-09/HL/NHLBI NIH HHS/United States
- R01HL072166/HL/NHLBI NIH HHS/United States
- R01 HL085629-03/HL/NHLBI NIH HHS/United States
- R01 HL085629-05/HL/NHLBI NIH HHS/United States
- R01 HL072166-06A1/HL/NHLBI NIH HHS/United States
- R01HL085629/HL/NHLBI NIH HHS/United States
- P20 RR015567/RR/NCRR NIH HHS/United States
- 5P20RR015567/RR/NCRR NIH HHS/United States
- R01 HL085629-04/HL/NHLBI NIH HHS/United States
- R01 HL085629-01/HL/NHLBI NIH HHS/United States
- R01 HL072166-07/HL/NHLBI NIH HHS/United States
- R01 HL072166/HL/NHLBI NIH HHS/United States
- R01 HL072166-08/HL/NHLBI NIH HHS/United States
- R01 HL085629/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

