Gonadotropin-releasing hormone pulse sensitivity of follicle-stimulating hormone-beta gene is mediated by differential expression of positive regulatory activator protein 1 factors and corepressors SKIL and TGIF1
- PMID: 21659477
- PMCID: PMC3146254
- DOI: 10.1210/me.2011-0032
Gonadotropin-releasing hormone pulse sensitivity of follicle-stimulating hormone-beta gene is mediated by differential expression of positive regulatory activator protein 1 factors and corepressors SKIL and TGIF1
Abstract
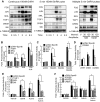
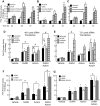
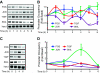
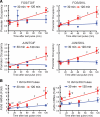
Gonadotropin synthesis and release is dependent on pulsatile stimulation by the hypothalamic neuropeptide GnRH. Generally, slow GnRH pulses promote FSH production, whereas rapid pulses favor LH, but the molecular mechanism underlying this pulse sensitivity is poorly understood. In this study, we developed and tested a model for FSHβ regulation in mouse LβT2 gonadotropes. By mining a previous microarray data set, we found that mRNA for positive regulators of Fshb expression, such as Fos and Jun, were up-regulated at slower pulse frequencies than a number of potential negative regulators, such as the corepressors Skil, Crem, and Tgif1. These latter corepressors reduced Fshb promoter activity whether driven by transfection of individual transcription factors or by treatment with GnRH and activin. Overexpression of binding or phosphorylation-defective ski-oncogene-like protein (SKIL) and TG interacting factor (TGIF1) mutants, however, failed to repress Fshb promoter activity. Knockdown of the endogenous repressors SKIL and TGIF1, but not cAMP response element-modulator, increased Fshb promoter activity driven by constant GnRH or activin. Chromatin immunoprecipitation analysis showed that FOS, SKIL, and TGIF1 occupy the FSHβ promoter in a cyclical manner after GnRH stimulation. Overexpression of corepressors SKIL or TGIF1 repressed induction of the Fshb promoter at the slow GnRH pulse frequency but had little effect at the fast pulse frequency. In contrast, knockdown of endogenous SKIL or TGIF1 selectively increased Fshb mRNA at the fast GnRH pulse frequency. Therefore, we propose a potential mechanism by which production of gonadotropin Fshb is modulated by positive transcription factors and negative corepressors with different pulse sensitivities.
Figures
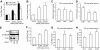

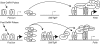
Similar articles
-
GnRH Pulse Frequency Control of Fshb Gene Expression Is Mediated via ERK1/2 Regulation of ICER.Mol Endocrinol. 2016 Mar;30(3):348-60. doi: 10.1210/me.2015-1222. Epub 2016 Feb 2. Mol Endocrinol. 2016. PMID: 26835742 Free PMC article.
-
Forkhead box O1 is a repressor of basal and GnRH-induced Fshb transcription in gonadotropes.Mol Endocrinol. 2013 Nov;27(11):1825-39. doi: 10.1210/me.2013-1185. Epub 2013 Sep 24. Mol Endocrinol. 2013. PMID: 24065703 Free PMC article.
-
Modeling and high-throughput experimental data uncover the mechanisms underlying Fshb gene sensitivity to gonadotropin-releasing hormone pulse frequency.J Biol Chem. 2017 Jun 9;292(23):9815-9829. doi: 10.1074/jbc.M117.783886. Epub 2017 Apr 6. J Biol Chem. 2017. PMID: 28385888 Free PMC article.
-
Possible role of PACAP and its PAC1 receptor in the differential regulation of pituitary LHbeta- and FSHbeta-subunit gene expression by pulsatile GnRH stimulation.Biol Reprod. 2013 Feb 14;88(2):35. doi: 10.1095/biolreprod.112.105601. Print 2013 Feb. Biol Reprod. 2013. PMID: 23197164 Review.
-
Regulation of gonadotropin subunit genes in tilapia.Comp Biochem Physiol B Biochem Mol Biol. 2001 Jun;129(2-3):489-502. doi: 10.1016/s1096-4959(01)00345-1. Comp Biochem Physiol B Biochem Mol Biol. 2001. PMID: 11399484 Review.
Cited by
-
Free fatty acids induce Lhb mRNA but suppress Fshb mRNA in pituitary LβT2 gonadotropes and diet-induced obesity reduces FSH levels in male mice and disrupts the proestrous LH/FSH surge in female mice.Endocrinology. 2013 Jun;154(6):2188-99. doi: 10.1210/en.2012-2218. Epub 2013 Mar 22. Endocrinology. 2013. PMID: 23525221 Free PMC article.
-
GnRH pulse frequency-dependent stimulation of FSHβ transcription is mediated via activation of PKA and CREB.Mol Endocrinol. 2013 Apr;27(4):606-18. doi: 10.1210/me.2012-1281. Epub 2013 Feb 7. Mol Endocrinol. 2013. PMID: 23393127 Free PMC article.
-
Outside the box signaling: secreted factors modulate GnRH receptor-mediated gonadotropin regulation.Mol Cell Endocrinol. 2014 Mar 25;385(1-2):56-61. doi: 10.1016/j.mce.2013.08.015. Epub 2013 Aug 28. Mol Cell Endocrinol. 2014. PMID: 23994024 Free PMC article. Review.
-
Ignored adult primary hypothyroidism presenting chiefly with persistent ovarian cysts: a need for increased awareness.Reprod Biol Endocrinol. 2011 Aug 23;9:119. doi: 10.1186/1477-7827-9-119. Reprod Biol Endocrinol. 2011. PMID: 21861901 Free PMC article. Review.
-
Regulation of reproduction via tight control of gonadotropin hormone levels.Mol Cell Endocrinol. 2018 Mar 5;463:116-130. doi: 10.1016/j.mce.2017.03.022. Epub 2017 Mar 22. Mol Cell Endocrinol. 2018. PMID: 28342855 Free PMC article. Review.
References
-
- Conn PM, Crowley WF., Jr 1994. Gonadotropin-releasing hormone and its analogs. Annu Rev Med 45:391–405 - PubMed
-
- Kaiser UB, Conn PM, Chin WW. 1997. Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocr Rev 18:46–70 - PubMed
-
- Lahlou N, Chabbert-Buffet N, Christin-Maitre S, Le Nestour E, Roger M, Bouchard P. 1999. Main inhibitor of follicle stimulating hormone in the luteal-follicular transition: inhibin A, oestradiol, or inhibin B? Hum Reprod 14:1190–1193 - PubMed
-
- Welt CK, Pagan YL, Smith PC, Rado KB, Hall JE. 2003. Control of follicle-stimulating hormone by estradiol and the inhibins: the critical role of estradiol at the hypothalamus during the luteal-follicular transition. J Clin Endocrinol Metab 88:1766–1771 - PubMed
-
- McDowell IF, Morris JF, Charlton HM. 1982. Characterization of the pituitary gonadotroph cells of hypogonadal (hpg) male mice: comparison with normal mice. J Endocrinol 95:321–330 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

