Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development
- PMID: 21651944
- PMCID: PMC3170448
- DOI: 10.1016/j.vaccine.2011.05.080
Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development
Abstract
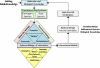
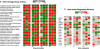
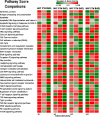
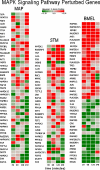
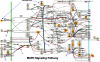
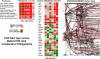
The aim of research on infectious diseases is their prevention, and brucellosis and salmonellosis as such are classic examples of worldwide zoonoses for application of a systems biology approach for enhanced rational vaccine development. When used optimally, vaccines prevent disease manifestations, reduce transmission of disease, decrease the need for pharmaceutical intervention, and improve the health and welfare of animals, as well as indirectly protecting against zoonotic diseases of people. Advances in the last decade or so using comprehensive systems biology approaches linking genomics, proteomics, bioinformatics, and biotechnology with immunology, pathogenesis and vaccine formulation and delivery are expected to enable enhanced approaches to vaccine development. The goal of this paper is to evaluate the role of computational systems biology analysis of host:pathogen interactions (the interactome) as a tool for enhanced rational design of vaccines. Systems biology is bringing a new, more robust approach to veterinary vaccine design based upon a deeper understanding of the host-pathogen interactions and its impact on the host's molecular network of the immune system. A computational systems biology method was utilized to create interactome models of the host responses to Brucella melitensis (BMEL), Mycobacterium avium paratuberculosis (MAP), Salmonella enterica Typhimurium (STM), and a Salmonella mutant (isogenic ΔsipA, sopABDE2) and linked to the basis for rational development of vaccines for brucellosis and salmonellosis as reviewed by Adams et al. and Ficht et al. [1,2]. A bovine ligated ileal loop biological model was established to capture the host gene expression response at multiple time points post infection. New methods based on Dynamic Bayesian Network (DBN) machine learning were employed to conduct a comparative pathogenicity analysis of 219 signaling and metabolic pathways and 1620 gene ontology (GO) categories that defined the host's biosignatures to each infectious condition. Through this DBN computational approach, the method identified significantly perturbed pathways and GO category groups of genes that define the pathogenicity signatures of the infectious agent. Our preliminary results provide deeper understanding of the overall complexity of host innate immune response as well as the identification of host gene perturbations that defines a unique host temporal biosignature response to each pathogen. The application of advanced computational methods for developing interactome models based on DBNs has proven to be instrumental in elucidating novel host responses and improved functional biological insight into the host defensive mechanisms. Evaluating the unique differences in pathway and GO perturbations across pathogen conditions allowed the identification of plausible host-pathogen interaction mechanisms. Accordingly, a systems biology approach to study molecular pathway gene expression profiles of host cellular responses to microbial pathogens holds great promise as a methodology to identify, model and predict the overall dynamics of the host-pathogen interactome. Thus, we propose that such an approach has immediate application to the rational design of brucellosis and salmonellosis vaccines.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Systems biology analysis of Brucella infected Peyer's patch reveals rapid invasion with modest transient perturbations of the host transcriptome.PLoS One. 2013 Dec 9;8(12):e81719. doi: 10.1371/journal.pone.0081719. eCollection 2013. PLoS One. 2013. PMID: 24349118 Free PMC article.
-
Multi-comparative systems biology analysis reveals time-course biosignatures of in vivo bovine pathway responses to B.melitensis, S.enterica Typhimurium and M.avium paratuberculosis.BMC Proc. 2011 Jun 3;5 Suppl 4(Suppl 4):S6. doi: 10.1186/1753-6561-5-S4-S6. BMC Proc. 2011. PMID: 21645321 Free PMC article.
-
Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach.PLoS One. 2011;6(11):e26869. doi: 10.1371/journal.pone.0026869. Epub 2011 Nov 11. PLoS One. 2011. PMID: 22096503 Free PMC article.
-
A rational framework for evaluating the next generation of vaccines against Mycobacterium avium subspecies paratuberculosis.Front Cell Infect Microbiol. 2014 Sep 9;4:126. doi: 10.3389/fcimb.2014.00126. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25250245 Free PMC article. Review.
-
Host-Brucella interactions and the Brucella genome as tools for subunit antigen discovery and immunization against brucellosis.Front Cell Infect Microbiol. 2013 May 16;3:17. doi: 10.3389/fcimb.2013.00017. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23720712 Free PMC article. Review.
Cited by
-
The characteristics and trend of adverse events following immunization reported by information system in Jiangsu province, China, 2015-2018.BMC Public Health. 2021 Jul 7;21(1):1338. doi: 10.1186/s12889-021-11387-3. BMC Public Health. 2021. PMID: 34229643 Free PMC article.
-
Systems biology analysis of Brucella infected Peyer's patch reveals rapid invasion with modest transient perturbations of the host transcriptome.PLoS One. 2013 Dec 9;8(12):e81719. doi: 10.1371/journal.pone.0081719. eCollection 2013. PLoS One. 2013. PMID: 24349118 Free PMC article.
-
Searching for animal models and potential target species for emerging pathogens: Experience gained from Middle East respiratory syndrome (MERS) coronavirus.One Health. 2017 Mar 3;3:34-40. doi: 10.1016/j.onehlt.2017.03.001. eCollection 2017 Jun. One Health. 2017. PMID: 28616501 Free PMC article. Review.
-
Translational research in infectious disease: current paradigms and challenges ahead.Transl Res. 2012 Jun;159(6):430-53. doi: 10.1016/j.trsl.2011.12.009. Epub 2012 Jan 15. Transl Res. 2012. PMID: 22633095 Free PMC article. Review.
-
Bridging the Gap Between Validation and Implementation of Non-Animal Veterinary Vaccine Potency Testing Methods.Animals (Basel). 2011 Nov 29;1(4):414-32. doi: 10.3390/ani1040414. Animals (Basel). 2011. PMID: 26486625 Free PMC article.
References
-
- Pacific Northwest National Laboratory (Mass Spec) http://www.pnl.gov/biology/programs/msd/measurements.stm.
-
- Adams LG, Babiuk Lorne, McGavin David, Nordgren Robert. Vaccines for Biodefense and Emerging Diseases. Academic Press; Amsterdam: 2009. Chapter 16. Special Issues Around Veterinary Vaccines; pp. 226–254. A. D. T. Barrett, Stanberry, Lawrence R.
-
- Ficht TA, Adams L. Garry. Vaccines for Biodefense and Emerging and Neglected Diseases. Academic Press; Amsterdam: 2009. Chapter 42. Brucellosis; pp. 808–829. A. D. T. Barrett, Stanberry, Lawrence R.
Publication types
MeSH terms
Substances
Grants and funding
- AI040124/AI/NIAID NIH HHS/United States
- U54 AI057156/AI/NIAID NIH HHS/United States
- R01 AI076246/AI/NIAID NIH HHS/United States
- R01 AI044170/AI/NIAID NIH HHS/United States
- AI060933/AI/NIAID NIH HHS/United States
- AI076246/AI/NIAID NIH HHS/United States
- R43AI084223-01/AI/NIAID NIH HHS/United States
- AI044170/AI/NIAID NIH HHS/United States
- R01 AI044170-10/AI/NIAID NIH HHS/United States
- AI079173/AI/NIAID NIH HHS/United States
- R21 AI079173/AI/NIAID NIH HHS/United States
- R43 AI084223/AI/NIAID NIH HHS/United States
- R29 AI040124/AI/NIAID NIH HHS/United States
- R44 AI058362/AI/NIAID NIH HHS/United States
- K08 AI060933/AI/NIAID NIH HHS/United States
- U54 AI057156,/AI/NIAID NIH HHS/United States
- R01 AI040124/AI/NIAID NIH HHS/United States
- 2R44AI058362-02/AI/NIAID NIH HHS/United States
- R01 AI044170-12/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical