Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway
- PMID: 21651802
- PMCID: PMC3127840
- DOI: 10.1186/1743-422X-8-285
Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway
Abstract
Background: Influenza viruses can modulate and hijack several cellular signalling pathways to efficiently support their replication. We recently investigated and compared the cellular gene expression profiles of human lung A549 cells infected by five different subtypes of human and avian influenza viruses (Josset et al. Plos One 2010). Using these transcriptomic data, we have focused our analysis on the modulation of the p53 pathway in response to influenza infection.
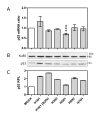
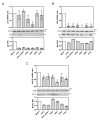
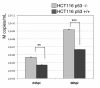
Results: Our results were supported by both RT-qPCR and western blot analyses and reveal multiple alterations of the p53 pathway during infection. A down-regulation of mRNA expression was observed for the main regulators of p53 protein stability during infection by the complete set of viruses tested, and a significant decrease in p53 mRNA expression was also observed in H5N1 infected cells. In addition, several p53 target genes were also down-regulated by these influenza viruses and the expression of their product reduced.
Conclusions: Our data reveal that influenza viruses cause an overall down-regulation of the host p53 pathway and highlight this pathway and p53 protein itself as important viral targets in the altering of apoptotic processes and in cell-cycle regulation.
Figures


Similar articles
-
Influenza A viruses control expression of proviral human p53 isoforms p53β and Delta133p53α.J Virol. 2012 Aug;86(16):8452-60. doi: 10.1128/JVI.07143-11. Epub 2012 May 30. J Virol. 2012. PMID: 22647703 Free PMC article.
-
The Nonstructural NS1 Protein of Influenza Viruses Modulates TP53 Splicing through Host Factor CPSF4.J Virol. 2019 Mar 21;93(7):e02168-18. doi: 10.1128/JVI.02168-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651364 Free PMC article.
-
Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a.J Gen Virol. 2013 May;94(Pt 5):985-995. doi: 10.1099/vir.0.049528-0. Epub 2013 Jan 23. J Gen Virol. 2013. PMID: 23343627
-
Evasion of influenza A viruses from innate and adaptive immune responses.Viruses. 2012 Sep;4(9):1438-76. doi: 10.3390/v4091438. Epub 2012 Sep 3. Viruses. 2012. PMID: 23170167 Free PMC article. Review.
-
p53 and RNA viruses: The tug of war.Wiley Interdiscip Rev RNA. 2023 Nov 20:e1826. doi: 10.1002/wrna.1826. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37985142 Review.
Cited by
-
Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein.J Virol. 2013 Mar;87(6):3039-52. doi: 10.1128/JVI.03176-12. Epub 2013 Jan 2. J Virol. 2013. PMID: 23283961 Free PMC article.
-
Influenza A viruses control expression of proviral human p53 isoforms p53β and Delta133p53α.J Virol. 2012 Aug;86(16):8452-60. doi: 10.1128/JVI.07143-11. Epub 2012 May 30. J Virol. 2012. PMID: 22647703 Free PMC article.
-
Magnetofluidic platform for rapid multiplexed screening of SARS-CoV-2 variants and respiratory pathogens.medRxiv [Preprint]. 2021 May 11:2021.05.10.21256995. doi: 10.1101/2021.05.10.21256995. medRxiv. 2021. Update in: Adv Mater Technol. 2022 Jun;7(6):2101013. doi: 10.1002/admt.202101013 PMID: 34013284 Free PMC article. Updated. Preprint.
-
The Nonstructural NS1 Protein of Influenza Viruses Modulates TP53 Splicing through Host Factor CPSF4.J Virol. 2019 Mar 21;93(7):e02168-18. doi: 10.1128/JVI.02168-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651364 Free PMC article.
-
Triggering Degradation of Host Cellular Proteins for Robust Propagation of Influenza Viruses.Int J Mol Sci. 2024 Apr 25;25(9):4677. doi: 10.3390/ijms25094677. Int J Mol Sci. 2024. PMID: 38731896 Free PMC article. Review.
References
-
- Palese P, Shaw M. In: Fields Virology. 5. Knipe D, Howley PM, editor. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. Orthomyxoviridae: the viruses and their replication; pp. 1647–1689.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

