Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors
- PMID: 21637844
- PMCID: PMC3102681
- DOI: 10.1371/journal.pone.0020197
Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors
Abstract
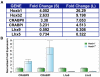
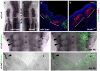
The evolutionarily conserved Hox family of homeodomain transcription factors plays fundamental roles in regulating cell specification along the anterior posterior axis during development of all bilaterian animals by controlling cell fate choices in a highly localized, extracellular signal and cell context dependent manner. Some studies have established downstream target genes in specific systems but their identification is insufficient to explain either the ability of Hox genes to direct homeotic transformations or the breadth of their patterning potential. To begin delineating Hox gene function in neural development we used a mouse ES cell based system that combines efficient neural differentiation with inducible Hoxb1 expression. Gene expression profiling suggested that Hoxb1 acted as both activator and repressor in the short term but predominantly as a repressor in the long run. Activated and repressed genes segregated in distinct processes suggesting that, in the context examined, Hoxb1 blocked differentiation while activating genes related to early developmental processes, wnt and cell surface receptor linked signal transduction and cell-to-cell communication. To further elucidate aspects of Hoxb1 function we used loss and gain of function approaches in the mouse and chick embryos. We show that Hoxb1 acts as an activator to establish the full expression domain of CRABPI and II in rhombomere 4 and as a repressor to restrict expression of Lhx5 and Lhx9. Thus the Hoxb1 patterning activity includes the regulation of the cellular response to retinoic acid and the delay of the expression of genes that commit cells to neural differentiation. The results of this study show that ES neural differentiation and inducible Hox gene expression can be used as a sensitive model system to systematically identify Hox novel target genes, delineate their interactions with signaling pathways in dictating cell fate and define the extent of functional overlap among different Hox genes.
Conflict of interest statement
Figures



Similar articles
-
Targeted germ line disruptions reveal general and species-specific roles for paralog group 1 hox genes in zebrafish.BMC Dev Biol. 2014 Jun 5;14:25. doi: 10.1186/1471-213X-14-25. BMC Dev Biol. 2014. PMID: 24902847 Free PMC article.
-
Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups.Development. 2002 Nov;129(22):5103-15. doi: 10.1242/dev.129.22.5103. Development. 2002. PMID: 12399303
-
Hoxb1 Regulates Distinct Signaling Pathways in Neuromesodermal and Hindbrain Progenitors to Promote Cell Survival and Specification.Stem Cells. 2022 Mar 16;40(2):175-189. doi: 10.1093/stmcls/sxab014. Stem Cells. 2022. PMID: 35257173
-
Hox genes in neural patterning and circuit formation in the mouse hindbrain.Curr Top Dev Biol. 2009;88:139-67. doi: 10.1016/S0070-2153(09)88005-8. Curr Top Dev Biol. 2009. PMID: 19651304 Review.
-
Hindbrain induction and patterning during early vertebrate development.Cell Mol Life Sci. 2019 Mar;76(5):941-960. doi: 10.1007/s00018-018-2974-x. Epub 2018 Dec 5. Cell Mol Life Sci. 2019. PMID: 30519881 Free PMC article. Review.
Cited by
-
Genetic insulin resistance is a potent regulator of gene expression and proliferation in human iPS cells.Diabetes. 2014 Dec;63(12):4130-42. doi: 10.2337/db14-0109. Epub 2014 Jul 24. Diabetes. 2014. PMID: 25059784 Free PMC article.
-
Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):5838-5845. doi: 10.1073/pnas.1610612114. Proc Natl Acad Sci U S A. 2017. PMID: 28584089 Free PMC article.
-
Hox-dependent coordination of mouse cardiac progenitor cell patterning and differentiation.Elife. 2020 Aug 17;9:e55124. doi: 10.7554/eLife.55124. Elife. 2020. PMID: 32804075 Free PMC article.
-
A Hox gene controls lateral line cell migration by regulating chemokine receptor expression downstream of Wnt signaling.Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16892-7. doi: 10.1073/pnas.1306282110. Epub 2013 Sep 30. Proc Natl Acad Sci U S A. 2013. PMID: 24082091 Free PMC article.
-
Analysis of novel caudal hindbrain genes reveals different regulatory logic for gene expression in rhombomere 4 versus 5/6 in embryonic zebrafish.Neural Dev. 2018 Jun 26;13(1):13. doi: 10.1186/s13064-018-0112-y. Neural Dev. 2018. PMID: 29945667 Free PMC article.
References
-
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, et al. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. - PubMed
-
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. - PubMed
-
- Wagmaister JA, Gleason JE, Eisenmann DM. Transcriptional upregulation of the C. elegans Hox gene lin-39 during vulval cell fate specification. Mech Dev. 2006;123:135–150. - PubMed
-
- Hueber SD, Bezdan D, Henz SR, Blank M, Wu H, et al. Comparative analysis of Hox downstream genes in Drosophila. Development. 2007;134:381–392. - PubMed
-
- Grienenberger A, Merabet S, Manak J, Iltis I, Fabre A, et al. Tgfbeta signaling acts on a Hox response element to confer specificity and diversity to Hox protein function. Development. 2003;130:5445–5455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

