Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness
- PMID: 21637832
- PMCID: PMC3102668
- DOI: 10.1371/journal.pone.0019774
Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness
Abstract
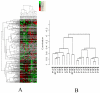
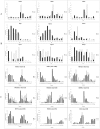
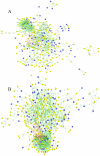
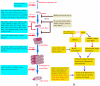
Understanding the dynamics of muscle transcriptome during development and between breeds differing in muscle growth is necessary to uncover the complex mechanism underlying muscle development. Herein, we present the first transcriptome-wide longissimus dorsi muscle development research concerning Lantang (LT, obese) and Landrace (LR, lean) pig breeds during 10 time-points from 35 days-post-coitus (dpc) to 180 days-post-natum (dpn) using Solexa/Illumina's Genome Analyzer. The data demonstrated that myogenesis was almost completed before 77 dpc, but the muscle phenotypes were still changed from 77 dpc to 28 dpn. Comparative analysis of the two breeds suggested that myogenesis started earlier but progressed more slowly in LT than in LR, the stages ranging from 49 dpc to 77 dpc are critical for formation of different muscle phenotypes. 595 differentially expressed myogenesis genes were identified, and their roles in myogenesis were discussed. Furthermore, GSK3B, IKBKB, ACVR1, ITGA and STMN1 might contribute to later myogenesis and more muscle fibers in LR than LT. Some myogenesis inhibitors (ID1, ID2, CABIN1, MSTN, SMAD4, CTNNA1, NOTCH2, GPC3 and HMOX1) were higher expressed in LT than in LR, which might contribute to more slow muscle differentiation in LT than in LR. We also identified several genes which might contribute to intramuscular adipose differentiation. Most important, we further proposed a novel model in which MyoD and MEF2A controls the balance between intramuscular adipogenesis and myogenesis by regulating CEBP family; Myf5 and MEF2C are essential during the whole myogenesis process while MEF2D affects muscle growth and maturation. The MRFs and MEF2 families are also critical for the phenotypic differences between the two pig breeds. Overall, this study contributes to elucidating the mechanism underlying muscle development, which could provide valuable information for pig meat quality improvement. The raw data have been submitted to Gene Expression Omnibus (GEO) under series GSE25406.
Conflict of interest statement
Figures




Similar articles
-
MicroRNA expression profiles differ between primary myofiber of lean and obese pig breeds.PLoS One. 2017 Jul 31;12(7):e0181897. doi: 10.1371/journal.pone.0181897. eCollection 2017. PLoS One. 2017. PMID: 28759650 Free PMC article.
-
Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity.BMC Dev Biol. 2007 Oct 1;7:109. doi: 10.1186/1471-213X-7-109. BMC Dev Biol. 2007. PMID: 17908293 Free PMC article.
-
LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs.Genome Biol. 2007;8(6):R115. doi: 10.1186/gb-2007-8-6-r115. Genome Biol. 2007. PMID: 17573972 Free PMC article.
-
Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.Semin Cell Dev Biol. 2017 Dec;72:19-32. doi: 10.1016/j.semcdb.2017.11.011. Epub 2017 Nov 15. Semin Cell Dev Biol. 2017. PMID: 29127046 Review.
-
[Interactions of proliferation and differentiation signaling pathways in myogenesis].Postepy Hig Med Dosw (Online). 2014 May 8;68:516-26. doi: 10.5604/17322693.1101617. Postepy Hig Med Dosw (Online). 2014. PMID: 24864103 Review. Polish.
Cited by
-
The Expression Pattern of p32 in Sheep Muscle and Its Role in Differentiation, Cell Proliferation, and Apoptosis of Myoblasts.Int J Mol Sci. 2019 Oct 18;20(20):5161. doi: 10.3390/ijms20205161. Int J Mol Sci. 2019. PMID: 31635221 Free PMC article.
-
Identification of circRNA-associated ceRNA networks using longissimus thoracis of pigs of different breeds and growth stages.BMC Genomics. 2022 Apr 11;23(1):294. doi: 10.1186/s12864-022-08515-7. BMC Genomics. 2022. PMID: 35410129 Free PMC article.
-
Transcript profile of skeletal muscle lipid metabolism genes affected by diet in a piglet model of low birth weight.PLoS One. 2019 Oct 29;14(10):e0224484. doi: 10.1371/journal.pone.0224484. eCollection 2019. PLoS One. 2019. PMID: 31661531 Free PMC article.
-
A new approach of gene co-expression network inference reveals significant biological processes involved in porcine muscle development in late gestation.Sci Rep. 2018 Jul 5;8(1):10150. doi: 10.1038/s41598-018-28173-8. Sci Rep. 2018. PMID: 29977047 Free PMC article.
-
Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics.BMC Genomics. 2014 May 31;15:413. doi: 10.1186/1471-2164-15-413. BMC Genomics. 2014. PMID: 24885501 Free PMC article.
References
-
- Li J, Chen Z, Liu D, Liu X, Sun B, et al. Genetic effects of IGF-1 gene on the performance in Landrace × Lantang pig resource population. J Genet Genomics. 2003;30:835–839. - PubMed
-
- Newcom DW, Stalder KJ, Baas TJ, Goodwin RN, Parrish FC, et al. Breed differences and genetic parameters of myoglobin concentration in porcine longissimus muscle. J Anim Sci. 2004;82:2264–2268. - PubMed
-
- Suzuki A, Kojima N, Ikeuchi Y, Ikarashi S, Moriyama N, et al. Carcass composition and meat quality of Chinese purebred and European × Chinese crossbred pigs. Meat Science. 1991;29:31–41. - PubMed
-
- Schook L, Beattie C, Beever J, Donovan S, Jamison R, et al. Swine in biomedical research: creating the building blocks of animal models. Anim Biotechnol. 2005;16:183–190. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

