Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine
- PMID: 21628671
- PMCID: PMC3143670
- DOI: 10.1093/infdis/jir199
Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine
Abstract
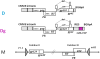
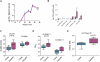
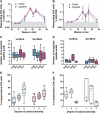
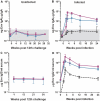
A simian immunodeficiency virus (SIV) vaccine coexpressing granulocyte-macrophage colony stimulating factor (GM-CSF) prevented infection in 71% of macaques that received 12 rectal challenges. The SIVsmE660 challenge had the tropism of incident human immunodeficiency virus (HIV) infections and a similar genetic distance from the SIV239 vaccine as intraclade HIV isolates. The heterologous prime-boost vaccine regimen used recombinant DNA for priming and recombinant modified vaccinia Ankara for boosting. Co-expression of GM-CSF in the DNA prime enhanced the avidity of elicited immunoglobulin G for SIV envelope glycoproteins, the titers of neutralizing antibody for easy-to-neutralize SIV isolates, and antibody-dependent cellular cytotoxicity. Impressively, the co-expressed GM-CSF increased vaccine-induced prevention of infection from 25% in the non-GM-CSF co-expressing vaccine group to 71% in the GM-CSF co-expressing vaccine group. The prevention of infection showed a strong correlation with the avidity of the elicited Env-specific antibody for the Env of the SIVsmE660 challenge virus (r = 0.9; P < .0001).
Figures

Similar articles
-
High Doses of GM-CSF Inhibit Antibody Responses in Rectal Secretions and Diminish Modified Vaccinia Ankara/Simian Immunodeficiency Virus Vaccine Protection in TRIM5α-Restrictive Macaques.J Immunol. 2016 Nov 1;197(9):3586-3596. doi: 10.4049/jimmunol.1600629. Epub 2016 Sep 28. J Immunol. 2016. PMID: 27683750 Free PMC article.
-
CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge.J Virol. 2014 Sep 1;88(17):9579-89. doi: 10.1128/JVI.00975-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920805 Free PMC article.
-
Virus-Like Particles Displaying Trimeric Simian Immunodeficiency Virus (SIV) Envelope gp160 Enhance the Breadth of DNA/Modified Vaccinia Virus Ankara SIV Vaccine-Induced Antibody Responses in Rhesus Macaques.J Virol. 2016 Sep 12;90(19):8842-54. doi: 10.1128/JVI.01163-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27466414 Free PMC article.
-
Co-expression of HIV-1 virus-like particles and granulocyte-macrophage colony stimulating factor by GEO-D03 DNA vaccine.Hum Vaccin Immunother. 2012 Nov 1;8(11):1654-8. doi: 10.4161/hv.21978. Epub 2012 Oct 30. Hum Vaccin Immunother. 2012. PMID: 23111169 Free PMC article. Review.
-
Immunogenicity and efficacy of DNA/MVA HIV vaccines in rhesus macaque models.Expert Rev Vaccines. 2017 Oct;16(10):973-985. doi: 10.1080/14760584.2017.1371594. Epub 2017 Sep 4. Expert Rev Vaccines. 2017. PMID: 28838267 Free PMC article. Review.
Cited by
-
Fcgbp - A Potential Viral Trap in RV144.Open AIDS J. 2014 Sep 8;8:21-4. doi: 10.2174/1874613601408010021. eCollection 2014. Open AIDS J. 2014. PMID: 25246998 Free PMC article.
-
Antigenicity, Immunogenicity and Protective Efficacy of Three Proteins Expressed in the Promastigote and Amastigote Stages of Leishmania infantum against Visceral Leishmaniasis.PLoS One. 2015 Sep 14;10(9):e0137683. doi: 10.1371/journal.pone.0137683. eCollection 2015. PLoS One. 2015. PMID: 26367128 Free PMC article.
-
Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques.J Virol. 2012 Aug;86(16):8835-47. doi: 10.1128/JVI.00923-12. Epub 2012 Jun 13. J Virol. 2012. PMID: 22696650 Free PMC article.
-
Oral Coadministration of an Intramuscular DNA/Modified Vaccinia Ankara Vaccine for Simian Immunodeficiency Virus Is Associated with Better Control of Infection in Orally Exposed Infant Macaques.AIDS Res Hum Retroviruses. 2019 Mar;35(3):310-325. doi: 10.1089/AID.2018.0180. Epub 2018 Nov 27. AIDS Res Hum Retroviruses. 2019. PMID: 30303405 Free PMC article.
-
Challenges in mucosal HIV vaccine development: lessons from non-human primate models.Viruses. 2014 Aug 15;6(8):3129-58. doi: 10.3390/v6083129. Viruses. 2014. PMID: 25196380 Free PMC article. Review.
References
-
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–0. - PubMed
-
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. - PubMed
-
- Borrello I, Pardoll D. GM-CSF-based cellular vaccines: a review of the clinical experience. Cytokine Growth Factor Rev. 2002;13:185–93. - PubMed
-
- Small EJ, Fong L. Developing immunotherapy as legitimate therapy for patients with prostate cancer. J Clin Oncol. 2010;28:1085–7. - PubMed
-
- Serafini P, Carbley R, Noonan KA, Tan G, Bronte V, Borrello I. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res. 2004;64:6337–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

