Exploiting nucleotide composition to engineer promoters
- PMID: 21625601
- PMCID: PMC3097237
- DOI: 10.1371/journal.pone.0020136
Exploiting nucleotide composition to engineer promoters
Abstract
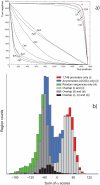
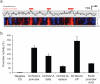
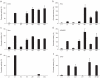
The choice of promoter is a critical step in optimizing the efficiency and stability of recombinant protein production in mammalian cell lines. Artificial promoters that provide stable expression across cell lines and can be designed to the desired strength constitute an alternative to the use of viral promoters. Here, we show how the nucleotide characteristics of highly active human promoters can be modelled via the genome-wide frequency distribution of short motifs: by overlapping motifs that occur infrequently in the genome, we constructed contiguous sequence that is rich in GC and CpGs, both features of known promoters, but lacking homology to real promoters. We show that snippets from this sequence, at 100 base pairs or longer, drive gene expression in vitro in a number of mammalian cells, and are thus candidates for use in protein production. We further show that expression is driven by the general transcription factors TFIIB and TFIID, both being ubiquitously present across cell types, which results in less tissue- and species-specific regulation compared to the viral promoter SV40. We lastly found that the strength of a promoter can be tuned up and down by modulating the counts of GC and CpGs in localized regions. These results constitute a "proof-of-concept" for custom-designing promoters that are suitable for biotechnological and medical applications.
Conflict of interest statement
Figures

Similar articles
-
Synthetic promoters for CHO cell engineering.Biotechnol Bioeng. 2014 Aug;111(8):1638-47. doi: 10.1002/bit.25227. Epub 2014 May 1. Biotechnol Bioeng. 2014. PMID: 24615264
-
Structure of vaccinia virus early promoters.J Mol Biol. 1989 Dec 20;210(4):749-69. doi: 10.1016/0022-2836(89)90107-1. J Mol Biol. 1989. PMID: 2515286
-
Interferon-gamma inhibits transgene expression driven by SV40 or CMV promoters but augments expression driven by the mammalian MHC I promoter.Hum Gene Ther. 1995 Oct;6(10):1291-7. doi: 10.1089/hum.1995.6.10-1291. Hum Gene Ther. 1995. PMID: 8590733
-
Identifying and engineering promoters for high level and sustainable therapeutic recombinant protein production in cultured mammalian cells.Biotechnol Lett. 2014 Aug;36(8):1569-79. doi: 10.1007/s10529-014-1523-4. Epub 2014 Apr 16. Biotechnol Lett. 2014. PMID: 24737078 Review.
-
Engineered and Natural Promoters and Chromatin-Modifying Elements for Recombinant Protein Expression in CHO Cells.Biotechnol J. 2018 Mar;13(3):e1700232. doi: 10.1002/biot.201700232. Epub 2017 Dec 7. Biotechnol J. 2018. PMID: 29145694 Review.
Cited by
-
The PLOS ONE synthetic biology collection: six years and counting.PLoS One. 2012;7(8):e43231. doi: 10.1371/journal.pone.0043231. Epub 2012 Aug 15. PLoS One. 2012. PMID: 22916228 Free PMC article. Review.
-
RNAi mechanisms in Huntington's disease therapy: siRNA versus shRNA.Transl Neurodegener. 2017 Nov 27;6:30. doi: 10.1186/s40035-017-0101-9. eCollection 2017. Transl Neurodegener. 2017. PMID: 29209494 Free PMC article. Review.
-
Transcriptomics analysis of salt stress tolerance in the roots of the mangrove Avicennia officinalis.Sci Rep. 2017 Aug 30;7(1):10031. doi: 10.1038/s41598-017-10730-2. Sci Rep. 2017. PMID: 28855698 Free PMC article.
-
Location dependence of the transcriptional response of a potential G-quadruplex in gene promoters under oxidative stress.Nucleic Acids Res. 2019 Jun 4;47(10):5049-5060. doi: 10.1093/nar/gkz207. Nucleic Acids Res. 2019. PMID: 30916339 Free PMC article.
-
Analysis of Simian Endogenous Retrovirus (SERV) Full-Length Proviruses in Old World Monkey Genomes.Genes (Basel). 2022 Jan 10;13(1):119. doi: 10.3390/genes13010119. Genes (Basel). 2022. PMID: 35052460 Free PMC article.
References
-
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nature Biotechnology. 2004;22:1393–1398. - PubMed
-
- Omasa T, Onitsuka M, Kim W-D. Cell engineering and cultivation of Chinese hamster ovary (CHO) cells. Curr Pharm Biotechnol. 2010;11:233–240. - PubMed
-
- Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–949. - PubMed
-
- Butler J, Kadonaga J. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

