OEP61 is a chaperone receptor at the plastid outer envelope
- PMID: 21612577
- PMCID: PMC5026492
- DOI: 10.1042/BJ20110448
OEP61 is a chaperone receptor at the plastid outer envelope
Abstract
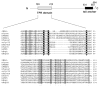
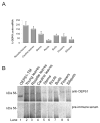
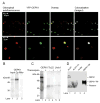
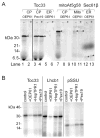
Chloroplast precursor proteins encoded in the nucleus depend on their targeting sequences for delivery to chloroplasts. There exist different routes to the chloroplast outer envelope, but a common theme is the involvement of molecular chaperones. Hsp90 (heat-shock protein 90) delivers precursors via its receptor Toc64, which transfers precursors to the core translocase in the outer envelope. In the present paper, we identify an uncharacterized protein in Arabidopsis thaliana OEP61 which shares common features with Toc64, and potentially provides an alternative route to the chloroplasts. Sequence analysis indicates that OEP61 possesses a clamp-type TPR (tetratricopeptide repeat) domain capable of binding molecular chaperones, and a C-terminal TMD (transmembrane domain). Phylogenetic comparisons show sequence similarities between the TPR domain of OEP61 and those of the Toc64 family. Expression of mRNA and protein was detected in all plant tissues, and localization at the chloroplast outer envelope was demonstrated by a combination of microscopy and in vitro import assays. Binding assays show that OEP61 interacts specifically with Hsp70 (heat-shock protein 70) via its TPR clamp domain. Furthermore, OEP61 selectively recognizes chloroplast precursors via their targeting sequences, and a soluble form of OEP61 inhibits chloroplast targeting. We therefore propose that OEP61 is a novel chaperone receptor at the chloroplast outer envelope, mediating Hsp70-dependent protein targeting to chloroplasts.
© The Authors Journal compilation © 2011 Biochemical Society
Figures


Similar articles
-
Exploring ligand recognition, selectivity and dynamics of TPR domains of chloroplast Toc64 and mitochondria Om64 from Arabidopsis thaliana.J Mol Recognit. 2014 Jun;27(6):402-14. doi: 10.1002/jmr.2360. J Mol Recognit. 2014. PMID: 24700626
-
A novel plastid-targeted J-domain protein in Arabidopsis thaliana.Plant Mol Biol. 2001 Jul;46(5):615-26. doi: 10.1023/a:1010665702621. Plant Mol Biol. 2001. PMID: 11516154
-
In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70.PLoS One. 2010 Sep 15;5(9):e12761. doi: 10.1371/journal.pone.0012761. PLoS One. 2010. PMID: 20856808 Free PMC article.
-
Structural components involved in plastid protein import.Essays Biochem. 2018 Apr 13;62(1):65-75. doi: 10.1042/EBC20170093. Print 2018 Apr 13. Essays Biochem. 2018. PMID: 29487196 Review.
-
Molecular chaperone involvement in chloroplast protein import.Biochim Biophys Acta. 2013 Feb;1833(2):332-40. doi: 10.1016/j.bbamcr.2012.03.019. Epub 2012 Apr 12. Biochim Biophys Acta. 2013. PMID: 22521451 Review.
Cited by
-
The Arabidopsis Plastidial Glucose-6-Phosphate Transporter GPT1 is Dually Targeted to Peroxisomes via the Endoplasmic Reticulum.Plant Cell. 2020 May;32(5):1703-1726. doi: 10.1105/tpc.19.00959. Epub 2020 Feb 28. Plant Cell. 2020. PMID: 32111666 Free PMC article.
-
The Plastid Outer Envelope - A Highly Dynamic Interface between Plastid and Cytoplasm.Front Plant Sci. 2011 Dec 14;2:97. doi: 10.3389/fpls.2011.00097. eCollection 2011. Front Plant Sci. 2011. PMID: 22629266 Free PMC article.
-
Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays.Front Plant Sci. 2012 Jun 28;3:148. doi: 10.3389/fpls.2012.00148. eCollection 2012. Front Plant Sci. 2012. PMID: 22783268 Free PMC article.
-
Quantification of interaction strengths between chaperones and tetratricopeptide repeat domain-containing membrane proteins.J Biol Chem. 2013 Oct 18;288(42):30614-30625. doi: 10.1074/jbc.M113.493015. Epub 2013 Sep 13. J Biol Chem. 2013. PMID: 24036116 Free PMC article.
-
The natural history of Get3-like chaperones.Traffic. 2019 May;20(5):311-324. doi: 10.1111/tra.12643. Traffic. 2019. PMID: 30972921 Free PMC article. Review.
References
-
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. - PubMed
-
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, Sohn EJ, Hwang I. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat Cell Biol. 2008;10:220–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

