The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks
- PMID: 21606185
- PMCID: PMC3147546
- DOI: 10.1128/IAI.05136-11
The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks
Abstract
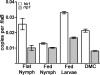
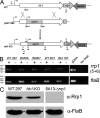
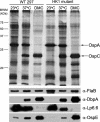
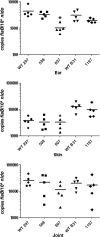
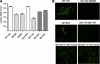
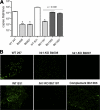
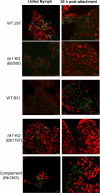
Two-component systems (TCS) are principal mechanisms by which bacteria adapt to their surroundings. Borrelia burgdorferi encodes only two TCS. One is comprised of a histidine kinase, Hk2, and the response regulator Rrp2. While the contribution of Hk2 remains unclear, Rrp2 is part of a regulatory pathway involving the spirochete's alternate sigma factors, RpoN and RpoS. Genes within the Rrp2/RpoN/RpoS regulon function to promote tick transmission and early infection. The other TCS consists of a hybrid histidine kinase, Hk1, and the response regulator Rrp1. Hk1 is composed of two periplasmic sensor domains (D1 and D2), followed by conserved cytoplasmic histidine kinase core, REC, and Hpt domains. In addition to its REC domain, Rrp1 contains a GGDEF motif characteristic of diguanylate cyclases. To investigate the role of Hk1 during the enzootic cycle, we inactivated this gene in two virulent backgrounds. Extensive characterization of the resulting mutants revealed a dramatic phenotype whereby Hk1-deficient spirochetes are virulent in mice and able to migrate out of the bite site during feeding but are killed within the midgut following acquisition. We hypothesize that the phosphorelay between Hk1 and Rrp1 is initiated by the binding of feeding-specific ligand(s) to Hk1 sensor domain D1 and/or D2. Once activated, Rrp1 directs the synthesis of cyclic dimeric GMP (c-di-GMP), which, in turn, modulates the expression and/or activity of gene products required for survival within feeding ticks. In contrast to the Rrp2/RpoN/RpoS pathway, which is active only within feeding nymphs, the Hk1/Rrp1 TCS is essential for survival during both larval and nymphal blood meals.
Figures

Similar articles
-
Structural characterization and modeling of the Borrelia burgdorferi hybrid histidine kinase Hk1 periplasmic sensor: A system for sensing small molecules associated with tick feeding.J Struct Biol. 2015 Oct;192(1):48-58. doi: 10.1016/j.jsb.2015.08.013. Epub 2015 Aug 28. J Struct Biol. 2015. PMID: 26321039 Free PMC article.
-
Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission.Infect Immun. 2015 Aug;83(8):3043-60. doi: 10.1128/IAI.00315-15. Epub 2015 May 18. Infect Immun. 2015. PMID: 25987708 Free PMC article.
-
The diguanylate cyclase, Rrp1, regulates critical steps in the enzootic cycle of the Lyme disease spirochetes.Mol Microbiol. 2011 Jul;81(1):219-31. doi: 10.1111/j.1365-2958.2011.07687.x. Epub 2011 Jun 5. Mol Microbiol. 2011. PMID: 21542866 Free PMC article.
-
Interaction of the Lyme disease spirochete with its tick vector.Cell Microbiol. 2016 Jul;18(7):919-27. doi: 10.1111/cmi.12609. Epub 2016 May 24. Cell Microbiol. 2016. PMID: 27147446 Free PMC article. Review.
-
Gene regulation in Borrelia burgdorferi.Annu Rev Microbiol. 2011;65:479-99. doi: 10.1146/annurev.micro.112408.134040. Annu Rev Microbiol. 2011. PMID: 21801026 Review.
Cited by
-
Motility is crucial for the infectious life cycle of Borrelia burgdorferi.Infect Immun. 2013 Jun;81(6):2012-21. doi: 10.1128/IAI.01228-12. Epub 2013 Mar 25. Infect Immun. 2013. PMID: 23529620 Free PMC article.
-
Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission.PLoS Pathog. 2012 Feb;8(2):e1002532. doi: 10.1371/journal.ppat.1002532. Epub 2012 Feb 16. PLoS Pathog. 2012. PMID: 22359504 Free PMC article.
-
The BB0345 Hypothetical Protein of Borrelia burgdorferi Is Essential for Mammalian Infection.Infect Immun. 2020 Nov 16;88(12):e00472-20. doi: 10.1128/IAI.00472-20. Print 2020 Nov 16. Infect Immun. 2020. PMID: 32928963 Free PMC article.
-
Two Different Virulence-Related Regulatory Pathways in Borrelia burgdorferi Are Directly Affected by Osmotic Fluxes in the Blood Meal of Feeding Ixodes Ticks.PLoS Pathog. 2016 Aug 15;12(8):e1005791. doi: 10.1371/journal.ppat.1005791. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27525653 Free PMC article.
-
Virulence of the Lyme disease spirochete before and after the tick bloodmeal: a quantitative assessment.Parasit Vectors. 2016 Mar 7;9:129. doi: 10.1186/s13071-016-1380-1. Parasit Vectors. 2016. PMID: 26951688 Free PMC article.
References
-
- Arnold K., Bordoli L., Kopp J., Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 - PubMed
-
- Balashov Y. S. 1972. Blood-sucking ticks (Ixodidae)-vectors of disease of man and animals. Misc. Pub. Entomol. Soc. Am. 8:161–376
-
- Bergstrom S., Zuckert W. R. 2010. Structure, function and biogenesis of the Borrelia cell envelope, p. 139–166 In Samuels D. S., Radolf J. D. (ed.), Borrelia: molecular biology, host interaction and pathogenesis. Calister Academic Press, Norfolk, United Kingdom
-
- Berntsson R. P. A., Smits S. H. J., Schmitt L., Slotboom D.-J., Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett. 584:2606–2617 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI059373/AI/NIAID NIH HHS/United States
- U54 AI057159/AI/NIAID NIH HHS/United States
- R03 AI085248/AI/NIAID NIH HHS/United States
- R21 AI085310/AI/NIAID NIH HHS/United States
- R01 AI059373/AI/NIAID NIH HHS/United States
- 3R01AI029735-20S1/AI/NIAID NIH HHS/United States
- U54 AI-057159/AI/NIAID NIH HHS/United States
- A1080615/PHS HHS/United States
- AI-29735/AI/NIAID NIH HHS/United States
- R56 AI029735/AI/NIAID NIH HHS/United States
- R01 AI029735/AI/NIAID NIH HHS/United States
- AI085310/AI/NIAID NIH HHS/United States
- AI059373/AI/NIAID NIH HHS/United States
- AI085248/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

