Culture-independent analysis of bacterial fuel contamination provides insight into the level of concordance with the standard industry practice of aerobic cultivation
- PMID: 21602386
- PMCID: PMC3127687
- DOI: 10.1128/AEM.02317-10
Culture-independent analysis of bacterial fuel contamination provides insight into the level of concordance with the standard industry practice of aerobic cultivation
Abstract
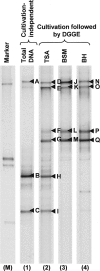
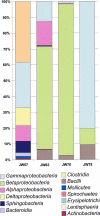
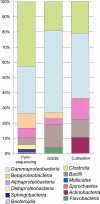
Bacterial diversity in contaminated fuels has not been systematically investigated using cultivation-independent methods. The fuel industry relies on phenotypic cultivation-based contaminant identification, which may lack accuracy and neglect difficult-to-culture taxa. By the use of industry practice aerobic cultivation, 16S rRNA gene sequencing, and strain genotyping, a collection of 152 unique contaminant isolates from 54 fuel samples was assembled, and a dominance of Pseudomonas (21%), Burkholderia (7%), and Bacillus (7%) was demonstrated. Denaturing gradient gel electrophoresis (DGGE) of 15 samples revealed Proteobacteria and Firmicutes to be the most abundant phyla. When 16S rRNA V6 gene pyrosequencing of four selected fuel samples (indicated by "JW") was performed, Betaproteobacteria (42.8%) and Gammaproteobacteria (30.6%) formed the largest proportion of reads; the most abundant genera were Marinobacter (15.4%; JW57), Achromobacter (41.6%; JW63), Burkholderia (80.7%; JW76), and Halomonas (66.2%; JW78), all of which were also observed by DGGE. However, the Clostridia (38.5%) and Deltaproteobacteria (11.1%) identified by pyrosequencing in sample JW57 were not observed by DGGE or aerobic culture. Genotyping revealed three instances where identical strains were found: (i) a Pseudomonas sp. strain recovered from 2 different diesel fuel tanks at a single industrial site; (ii) a Mangroveibacter sp. strain isolated from 3 biodiesel tanks at a single refinery site; and (iii) a Burkholderia vietnamiensis strain present in two unrelated automotive diesel samples. Overall, aerobic cultivation of fuel contaminants recovered isolates broadly representative of the phyla and classes present but lacked accuracy by overrepresenting members of certain groups such as Pseudomonas.
Figures



Similar articles
-
Microbial diversity in an Armenian geothermal spring assessed by molecular and culture-based methods.J Basic Microbiol. 2014 Nov;54(11):1240-50. doi: 10.1002/jobm.201300999. Epub 2014 Apr 16. J Basic Microbiol. 2014. PMID: 24740751
-
Comparative approach to capture bacterial diversity of coastal waters.J Microbiol. 2011 Oct;49(5):729-40. doi: 10.1007/s12275-011-1205-z. Epub 2011 Nov 9. J Microbiol. 2011. PMID: 22068488
-
The presence of embedded bacterial pure cultures in agar plates stimulate the culturability of soil bacteria.J Microbiol Methods. 2009 Nov;79(2):166-73. doi: 10.1016/j.mimet.2009.08.006. Epub 2009 Aug 20. J Microbiol Methods. 2009. PMID: 19699242
-
Gammaproteobacteria occurrence and microdiversity in Tyrrhenian Sea sediments as revealed by cultivation-dependent and -independent approaches.Syst Appl Microbiol. 2010 Jun;33(4):222-31. doi: 10.1016/j.syapm.2010.02.005. Epub 2010 Apr 22. Syst Appl Microbiol. 2010. PMID: 20413241
-
Molecular approaches: advantages and artifacts in assessing bacterial diversity.J Ind Microbiol Biotechnol. 2007 Jul;34(7):463-73. doi: 10.1007/s10295-007-0219-3. Epub 2007 May 3. J Ind Microbiol Biotechnol. 2007. PMID: 17476541 Review.
Cited by
-
A New Approach of Rpf Addition to Explore Bacterial Consortium for Enhanced Phenol Degradation Under High Salinity Conditions.Curr Microbiol. 2018 Aug;75(8):1046-1054. doi: 10.1007/s00284-018-1489-x. Epub 2018 Apr 6. Curr Microbiol. 2018. PMID: 29626248
-
Key role for efflux in the preservative susceptibility and adaptive resistance of Burkholderia cepacia complex bacteria.Antimicrob Agents Chemother. 2013 Jul;57(7):2972-80. doi: 10.1128/AAC.00140-13. Epub 2013 Apr 15. Antimicrob Agents Chemother. 2013. PMID: 23587949 Free PMC article.
-
Not all Pseudomonas aeruginosa are equal: strains from industrial sources possess uniquely large multireplicon genomes.Microb Genom. 2019 Jul;5(7):e000276. doi: 10.1099/mgen.0.000276. Epub 2019 Jun 6. Microb Genom. 2019. PMID: 31170060 Free PMC article.
-
Understanding the challenges of non-food industrial product contamination.FEMS Microbiol Lett. 2019 Dec 1;366(23):fnaa010. doi: 10.1093/femsle/fnaa010. FEMS Microbiol Lett. 2019. PMID: 31977006 Free PMC article. Review.
-
Quantifying bacterial efflux within subcellular domains of Pseudomonas aeruginosa.Appl Environ Microbiol. 2024 Nov 20;90(11):e0144724. doi: 10.1128/aem.01447-24. Epub 2024 Oct 30. Appl Environ Microbiol. 2024. PMID: 39475289 Free PMC article.
References
-
- Allsopp D., Seal K., Gaylarde C. C. 2004. Introduction to biodeterioration, 2nd ed. Cambridge University Press, Cambridge, United Kingdom
-
- Biddick R., Spilker T., Martin A., LiPuma J. J. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228:57–62 - PubMed
-
- Brown L. M., et al. 2010. Community dynamics and phylogenetics of bacteria fouling Jet A and JP-8 aviation fuel. Int. Biodeterior. Biodeg. 64:253–261
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

