Hierarchies in eukaryotic genome organization: Insights from polymer theory and simulations
- PMID: 21595865
- PMCID: PMC3102647
- DOI: 10.1186/2046-1682-4-8
Hierarchies in eukaryotic genome organization: Insights from polymer theory and simulations
Abstract
Eukaryotic genomes possess an elaborate and dynamic higher-order structure within the limiting confines of the cell nucleus. Knowledge of the physical principles and the molecular machinery that govern the 3D organization of this structure and its regulation are key to understanding the relationship between genome structure and function. Elegant microscopy and chromosome conformation capture techniques supported by analysis based on polymer models are important steps in this direction. Here, we review results from these efforts and provide some additional insights that elucidate the relationship between structure and function at different hierarchical levels of genome organization.
Figures


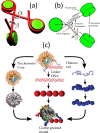
 and
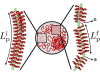
and  refer to the initial and final persistence length, respectively.
refer to the initial and final persistence length, respectively.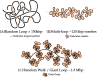


Similar articles
-
Plant 3D genomics: the exploration and application of chromatin organization.New Phytol. 2021 Jun;230(5):1772-1786. doi: 10.1111/nph.17262. Epub 2021 Mar 4. New Phytol. 2021. PMID: 33560539 Free PMC article. Review.
-
Computational models of large-scale genome architecture.Int Rev Cell Mol Biol. 2014;307:275-349. doi: 10.1016/B978-0-12-800046-5.00009-6. Int Rev Cell Mol Biol. 2014. PMID: 24380598 Review.
-
Bridging the resolution gap in structural modeling of 3D genome organization.PLoS Comput Biol. 2011 Jul;7(7):e1002125. doi: 10.1371/journal.pcbi.1002125. Epub 2011 Jul 14. PLoS Comput Biol. 2011. PMID: 21779160 Free PMC article. Review.
-
Computational methods for predicting 3D genomic organization from high-resolution chromosome conformation capture data.Brief Funct Genomics. 2020 Jul 29;19(4):292-308. doi: 10.1093/bfgp/elaa004. Brief Funct Genomics. 2020. PMID: 32353112 Free PMC article.
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
Cited by
-
Inferring 3D chromatin structure using a multiscale approach based on quaternions.BMC Bioinformatics. 2015 Jul 29;16:234. doi: 10.1186/s12859-015-0667-0. BMC Bioinformatics. 2015. PMID: 26220581 Free PMC article.
-
Mesoscale Modeling Reveals Hierarchical Looping of Chromatin Fibers Near Gene Regulatory Elements.J Phys Chem B. 2016 Aug 25;120(33):8642-53. doi: 10.1021/acs.jpcb.6b03197. Epub 2016 Jun 16. J Phys Chem B. 2016. PMID: 27218881 Free PMC article.
-
Recovering ensembles of chromatin conformations from contact probabilities.Nucleic Acids Res. 2013 Jan 7;41(1):63-75. doi: 10.1093/nar/gks1029. Epub 2012 Nov 11. Nucleic Acids Res. 2013. PMID: 23143266 Free PMC article.
-
Genome-wide profiling of chromosome interactions in Plasmodium falciparum characterizes nuclear architecture and reconfigurations associated with antigenic variation.Mol Microbiol. 2013 Nov;90(3):519-37. doi: 10.1111/mmi.12381. Epub 2013 Sep 30. Mol Microbiol. 2013. PMID: 23980881 Free PMC article.
-
Evaluation of novel design strategies for developing zinc finger nucleases tools for treating human diseases.Biotechnol Res Int. 2014;2014:970595. doi: 10.1155/2014/970595. Epub 2014 Apr 6. Biotechnol Res Int. 2014. PMID: 24808958 Free PMC article.

