Why eukaryotic cells use introns to enhance gene expression: splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity
- PMID: 21592350
- PMCID: PMC3118952
- DOI: 10.1186/1745-6150-6-24
Why eukaryotic cells use introns to enhance gene expression: splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity
Abstract
Background: The costs and benefits of spliceosomal introns in eukaryotes have not been established. One recognized effect of intron splicing is its known enhancement of gene expression. However, the mechanism regulating such splicing-mediated expression enhancement has not been defined. Previous studies have shown that intron splicing is a time-consuming process, indicating that splicing may not reduce the time required for transcription and processing of spliced pre-mRNA molecules; rather, it might facilitate the later rounds of transcription. Because the densities of active RNA polymerase II on most genes are less than one molecule per gene, direct interactions between the splicing apparatus and transcriptional complexes (from the later rounds of transcription) are infrequent, and thus unlikely to account for splicing-mediated gene expression enhancement.
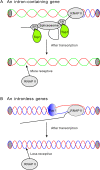
Presentation of the hypothesis: The serine/arginine-rich protein SF2/ASF can inhibit the DNA topoisomerase I activity that removes negative supercoiling of DNA generated by transcription. Consequently, splicing could make genes more receptive to RNA polymerase II during the later rounds of transcription, and thus affect the frequency of gene transcription. Compared with the transcriptional enhancement mediated by strong promoters, intron-containing genes experience a lower frequency of cut-and-paste processes. The cleavage and religation activity of DNA strands by DNA topoisomerase I was recently shown to account for transcription-associated mutagenesis. Therefore, intron-mediated enhancement of gene expression could reduce transcription-associated genome instability.
Testing the hypothesis: Experimentally test whether transcription-associated mutagenesis is lower in intron-containing genes than in intronless genes. Use bioinformatic analysis to check whether exons flanking lost introns have higher frequencies of short deletions.
Implications of the hypothesis: The mechanism of intron-mediated enhancement proposed here may also explain the positive correlation observed between intron size and gene expression levels in unicellular organisms, and the greater number of intron containing genes in higher organisms.
Reviewers: This article was reviewed by Dr Arcady Mushegian, Dr Igor B Rogozin (nominated by Dr I King Jordan) and Dr Alexey S Kondrashov. For the full reviews, please go to the Reviewer's Reports section.
Figures
Similar articles
-
Exon definition as a potential negative force against intron losses in evolution.Biol Direct. 2008 Nov 13;3:46. doi: 10.1186/1745-6150-3-46. Biol Direct. 2008. PMID: 19014515 Free PMC article.
-
Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence.Aging Cell. 2011 Oct;10(5):896-907. doi: 10.1111/j.1474-9726.2011.00727.x. Epub 2011 Jul 19. Aging Cell. 2011. PMID: 21668763
-
Mapping the SF2/ASF binding sites in the bovine growth hormone exonic splicing enhancer.J Biol Chem. 2000 Sep 15;275(37):29170-7. doi: 10.1074/jbc.M001126200. J Biol Chem. 2000. PMID: 10880506
-
How introns enhance gene expression.Int J Biochem Cell Biol. 2017 Oct;91(Pt B):145-155. doi: 10.1016/j.biocel.2017.06.016. Epub 2017 Jul 1. Int J Biochem Cell Biol. 2017. PMID: 28673892 Review.
-
Spliceosomal Introns: Features, Functions, and Evolution.Biochemistry (Mosc). 2020 Jul;85(7):725-734. doi: 10.1134/S0006297920070019. Biochemistry (Mosc). 2020. PMID: 33040717 Review.
Cited by
-
A Comparative Analysis of the Chloroplast Genomes of Four Polygonum Medicinal Plants.Front Genet. 2022 Apr 25;13:764534. doi: 10.3389/fgene.2022.764534. eCollection 2022. Front Genet. 2022. PMID: 35547259 Free PMC article.
-
The regulatory genome of the malaria vector Anopheles gambiae: integrating chromatin accessibility and gene expression.NAR Genom Bioinform. 2021 Jan 20;3(1):lqaa113. doi: 10.1093/nargab/lqaa113. eCollection 2021 Mar. NAR Genom Bioinform. 2021. PMID: 33987532 Free PMC article.
-
The Complete Plastid Genome of Magnolia zenii and Genetic Comparison to Magnoliaceae species.Molecules. 2019 Jan 11;24(2):261. doi: 10.3390/molecules24020261. Molecules. 2019. PMID: 30641990 Free PMC article.
-
Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops.BMC Genomics. 2015 Sep 21;16(1):721. doi: 10.1186/s12864-015-1914-5. BMC Genomics. 2015. PMID: 26391769 Free PMC article.
-
Characterization of the complete chloroplast genome of Brassica oleracea var. italica and phylogenetic relationships in Brassicaceae.PLoS One. 2022 Feb 24;17(2):e0263310. doi: 10.1371/journal.pone.0263310. eCollection 2022. PLoS One. 2022. PMID: 35202392 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

