Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity
- PMID: 21576369
- PMCID: PMC3133407
- DOI: 10.1128/MCB.00974-10
Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity
Abstract
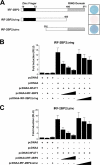
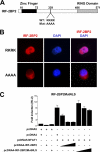
The nuclear factor of activated T cells (NFAT) family of transcription factors is expressed in a wide range of cell types and regulates genes involved in cell cycle, differentiation, and apoptosis. NFAT proteins share two well-conserved regions, the regulatory domain and the DNA binding domain. The N- and C-terminal ends are transactivation sites and show less sequence similarity, whereas their molecular functions remain poorly understood. Here, we identified a transcriptional repressor, interferon regulatory factor 2 binding protein 2 (IRF-2BP2), which specifically interacts with the C-terminal domain of NFAT1 among the NFAT family members. IRF-2BP2 was described as a corepressor by inhibiting both enhancer-activated and basal transcription. Gene reporter assays demonstrated that IRF-2BP2 represses the NFAT1-dependent transactivation of NFAT-responsive promoters. The ectopic expression of IRF-2BP2 in CD4 T cells resulted in decreased interleukin-2 (IL-2) and IL-4 production, supporting a repressive function of IRF-2BP2 for NFAT target genes. Furthermore, NFAT1 and IRF-2BP2 colocalized in the nucleus in activated cells, and the mutation of a newly identified nuclear localization signal in the IRF-2BP2 rendered it cytoplasmic, abolishing its repressive effect on NFAT1 activity. Collectively, our data demonstrate that IRF-2BP2 is a negative regulator of the NFAT1 transcription factor and suggest that NFAT1 repression occurs at the transcriptional level.
Figures


Similar articles
-
Identification of novel co-repressor molecules for Interferon Regulatory Factor-2.Nucleic Acids Res. 2003 Jun 15;31(12):3016-26. doi: 10.1093/nar/gkg431. Nucleic Acids Res. 2003. PMID: 12799427 Free PMC article.
-
Transcriptional regulation of the c-Myc promoter by NFAT1 involves negative and positive NFAT-responsive elements.Cell Cycle. 2012 Mar 1;11(5):1014-28. doi: 10.4161/cc.11.5.19518. Epub 2012 Mar 1. Cell Cycle. 2012. PMID: 22333584
-
T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells.J Immunol. 1999 Jan 15;162(2):860-70. J Immunol. 1999. PMID: 9916709
-
Nuclear factor of activated T cells 1 (NFAT1)-induced permissive chromatin modification facilitates nuclear factor-κB (NF-κB)-mediated interleukin-9 (IL-9) transactivation.J Biol Chem. 2012 May 4;287(19):15445-57. doi: 10.1074/jbc.M112.340356. Epub 2012 Mar 15. J Biol Chem. 2012. PMID: 22427656 Free PMC article.
-
Cell cycle and apoptosis regulation by NFAT transcription factors: new roles for an old player.Cell Death Dis. 2016 Apr 21;7(4):e2199. doi: 10.1038/cddis.2016.97. Cell Death Dis. 2016. PMID: 27100893 Free PMC article. Review.
Cited by
-
Novel frameshift variants expand the map of the genetic defects in IRF2BP2.Front Immunol. 2023 Oct 9;14:1279171. doi: 10.3389/fimmu.2023.1279171. eCollection 2023. Front Immunol. 2023. PMID: 37876937 Free PMC article.
-
IRF2BP2: A New Player at the Crossroads of Inflammation and Lipid Metabolism.Circ Res. 2015 Sep 25;117(8):656-8. doi: 10.1161/CIRCRESAHA.115.307245. Circ Res. 2015. PMID: 26405180 Free PMC article. No abstract available.
-
Control of developmentally primed erythroid genes by combinatorial co-repressor actions.Nat Commun. 2015 Nov 23;6:8893. doi: 10.1038/ncomms9893. Nat Commun. 2015. PMID: 26593974 Free PMC article.
-
IRF2BP2 transcriptional repressor restrains naive CD4 T cell activation and clonal expansion induced by TCR triggering.J Leukoc Biol. 2016 Nov;100(5):1081-1091. doi: 10.1189/jlb.2A0815-368R. Epub 2016 Jun 10. J Leukoc Biol. 2016. PMID: 27286791 Free PMC article.
-
The NFAT3/RERG Complex in Luminal Breast Cancers Is Required to Inhibit Cell Invasion and May Be Correlated With an Absence of Axillary Lymph Nodes Colonization.Front Oncol. 2022 Jun 30;12:804868. doi: 10.3389/fonc.2022.804868. eCollection 2022. Front Oncol. 2022. PMID: 35847954 Free PMC article.
References
-
- Baksh S., et al. 2002. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10:1071–1081 - PubMed
-
- Bartel P. L., Fields S. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241–263 - PubMed
-
- Bert A. G., Burrows J., Hawwari A., Vadas M. A., Cockerill P. N. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646–5655 - PubMed
-
- Bodor J., Bodorova J., Gress R. E. 2000. Suppression of T cell function: a potential role for transcriptional repressor ICER. J. Leukoc. Biol. 67:774–779 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
