Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier
- PMID: 21576223
- PMCID: PMC3159475
- DOI: 10.1093/nar/gkr340
Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier
Abstract
Most DNA double-strand breaks (DSBs) in S- and G2-phase cells are repaired accurately by Rad51-dependent sister chromatid recombination. However, a minority give rise to gross chromosome rearrangements (GCRs), which can result in disease/death. What determines whether a DSB is repaired accurately or inaccurately is currently unclear. We provide evidence that suggests that perturbing replication by a non-programmed protein-DNA replication fork barrier results in the persistence of replication intermediates (most likely regions of unreplicated DNA) into mitosis, which results in anaphase bridge formation and ultimately to DNA breakage. However, unlike previously characterised replication-associated DSBs, these breaks are repaired mainly by Rad51-independent processes such as single-strand annealing, and are therefore prone to generate GCRs. These data highlight how a replication-associated DSB can be predisposed to give rise to genome rearrangements in eukaryotes.
Figures
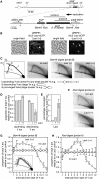
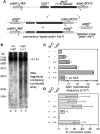
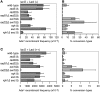
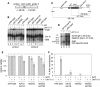
Similar articles
-
The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability.Genes Dev. 2012 Mar 15;26(6):594-602. doi: 10.1101/gad.184663.111. Genes Dev. 2012. PMID: 22426535 Free PMC article.
-
Schizosaccharomyces pombe Assays to Study Mitotic Recombination Outcomes.Genes (Basel). 2020 Jan 10;11(1):79. doi: 10.3390/genes11010079. Genes (Basel). 2020. PMID: 31936815 Free PMC article. Review.
-
Unprotected Replication Forks Are Converted into Mitotic Sister Chromatid Bridges.Mol Cell. 2017 May 4;66(3):398-410.e4. doi: 10.1016/j.molcel.2017.04.002. Mol Cell. 2017. PMID: 28475874
-
A new role for Rrm3 in repair of replication-born DNA breakage by sister chromatid recombination.PLoS Genet. 2017 May 5;13(5):e1006781. doi: 10.1371/journal.pgen.1006781. eCollection 2017 May. PLoS Genet. 2017. PMID: 28475600 Free PMC article.
-
A new class of ultrafine anaphase bridges generated by homologous recombination.Cell Cycle. 2018;17(17):2101-2109. doi: 10.1080/15384101.2018.1515555. Epub 2018 Sep 25. Cell Cycle. 2018. PMID: 30253678 Free PMC article. Review.
Cited by
-
The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability.Genes Dev. 2012 Mar 15;26(6):594-602. doi: 10.1101/gad.184663.111. Genes Dev. 2012. PMID: 22426535 Free PMC article.
-
The mechanism of DNA replication termination in vertebrates.Nature. 2015 Sep 17;525(7569):345-50. doi: 10.1038/nature14887. Epub 2015 Aug 31. Nature. 2015. PMID: 26322582 Free PMC article.
-
MHF1-2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination.Open Biol. 2013 Sep 11;3(9):130102. doi: 10.1098/rsob.130102. Open Biol. 2013. Retraction in: Open Biol. 2018 Feb;8(2):180010. doi: 10.1098/rsob.180010. PMID: 24026537 Free PMC article. Retracted.
-
Schizosaccharomyces pombe Assays to Study Mitotic Recombination Outcomes.Genes (Basel). 2020 Jan 10;11(1):79. doi: 10.3390/genes11010079. Genes (Basel). 2020. PMID: 31936815 Free PMC article. Review.
-
Immediate visualization of recombination events and chromosome segregation defects in fission yeast meiosis.Chromosoma. 2019 Sep;128(3):385-396. doi: 10.1007/s00412-019-00691-y. Epub 2019 Feb 9. Chromosoma. 2019. PMID: 30739171 Free PMC article.
References
-
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 2002;3:859–870. - PubMed
-
- Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000;14:1–10. - PubMed
-
- Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair. 2007;6:994–1003. - PubMed
-
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell. Biol. 2010;11:208–219. - PubMed
-
- Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell. 2010;39:346–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

