Metabolite-enabled eradication of bacterial persisters by aminoglycosides
- PMID: 21562562
- PMCID: PMC3145328
- DOI: 10.1038/nature10069
Metabolite-enabled eradication of bacterial persisters by aminoglycosides
Abstract
Bacterial persistence is a state in which a sub-population of dormant cells, or 'persisters', tolerates antibiotic treatment. Bacterial persisters have been implicated in biofilms and in chronic and recurrent infections. Despite this clinical relevance, there are currently no viable means for eradicating persisters. Here we show that specific metabolic stimuli enable the killing of both Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) persisters with aminoglycosides. This potentiation is aminoglycoside-specific, it does not rely on growth resumption and it is effective in both aerobic and anaerobic conditions. It proceeds by the generation of a proton-motive force which facilitates aminoglycoside uptake. Our results demonstrate that persisters, although dormant, are primed for metabolite uptake, central metabolism and respiration. We show that aminoglycosides can be used in combination with specific metabolites to treat E. coli and S. aureus biofilms. Furthermore, we demonstrate that this approach can improve the treatment of chronic infections in a mouse urinary tract infection model. This work establishes a strategy for eradicating bacterial persisters that is based on metabolism, and highlights the importance of the metabolic environment to antibiotic treatment.
Figures
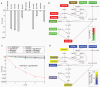
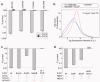
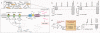

Comment in
-
Antimicrobials: Killing persisters while they sleep.Nat Rev Microbiol. 2011 Jun 16;9(7):482-3. doi: 10.1038/nrmicro2601. Nat Rev Microbiol. 2011. PMID: 21677681 No abstract available.
Similar articles
-
Rapid Freezing Enables Aminoglycosides To Eradicate Bacterial Persisters via Enhancing Mechanosensitive Channel MscL-Mediated Antibiotic Uptake.mBio. 2020 Feb 11;11(1):e03239-19. doi: 10.1128/mBio.03239-19. mBio. 2020. PMID: 32047133 Free PMC article.
-
pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms.J Infect Dis. 2014 Nov 1;210(9):1357-66. doi: 10.1093/infdis/jiu286. Epub 2014 May 15. J Infect Dis. 2014. PMID: 24837402
-
Hypoionic shock treatment enables aminoglycosides antibiotics to eradicate bacterial persisters.Sci Rep. 2015 Oct 5;5:14247. doi: 10.1038/srep14247. Sci Rep. 2015. PMID: 26435063 Free PMC article.
-
Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease.Bioessays. 2014 Oct;36(10):991-6. doi: 10.1002/bies.201400080. Epub 2014 Aug 6. Bioessays. 2014. PMID: 25100240 Review.
-
Multidrug tolerance of biofilms and persister cells.Curr Top Microbiol Immunol. 2008;322:107-31. doi: 10.1007/978-3-540-75418-3_6. Curr Top Microbiol Immunol. 2008. PMID: 18453274 Review.
Cited by
-
Rapid clinical bacteriology and its future impact.Ann Lab Med. 2013 Jan;33(1):14-27. doi: 10.3343/alm.2013.33.1.14. Epub 2012 Dec 17. Ann Lab Med. 2013. PMID: 23301218 Free PMC article. Review.
-
Persisters-as elusive as ever.Appl Microbiol Biotechnol. 2016 Aug;100(15):6545-6553. doi: 10.1007/s00253-016-7648-8. Epub 2016 Jun 4. Appl Microbiol Biotechnol. 2016. PMID: 27262568 Free PMC article. Review.
-
Effects of Helicobacter pylori eradication on the profiles of blood metabolites and their associations with the progression of gastric lesions: a prospective follow-up study.Cancer Biol Med. 2022 Aug 30;19(8):1259-73. doi: 10.20892/j.issn.2095-3941.2022.0255. Cancer Biol Med. 2022. PMID: 36069529 Free PMC article.
-
A method for high throughput determination of viable bacteria cell counts in 96-well plates.BMC Microbiol. 2012 Nov 13;12:259. doi: 10.1186/1471-2180-12-259. BMC Microbiol. 2012. PMID: 23148795 Free PMC article.
-
Gentamicin Combined With Hypoionic Shock Rapidly Eradicates Aquaculture Bacteria in vitro and in vivo.Front Microbiol. 2021 Apr 6;12:641846. doi: 10.3389/fmicb.2021.641846. eCollection 2021. Front Microbiol. 2021. PMID: 33889141 Free PMC article.
References
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. - PubMed
-
- Gefen O, Balaban NQ. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev. 2009;33:704–717. - PubMed
-
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. - PubMed
-
- Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem Biol. 2007;3:549–556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

