Dancing on DNA: kinetic aspects of search processes on DNA
- PMID: 21560221
- PMCID: PMC4590286
- DOI: 10.1002/cphc.201100112
Dancing on DNA: kinetic aspects of search processes on DNA
Abstract
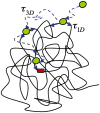
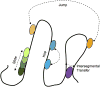
Recognition and binding of specific sites on DNA by proteins is central for many cellular functions such as transcription, replication, and recombination. In the search for its target site, the DNA-associated protein is facing both thermodynamic and kinetic difficulties. The thermodynamic challenge lies in recognizing and tightly binding a cognate (specific) site among the billions of other (non-specific) sequences on the DNA. The kinetic difficulty lies in finding a cognate site in mere seconds amidst the crowded cellular environment that is filled with other DNA sequences and proteins. Herein, we discuss the history of the DNA search problem, the theoretical background and the various experimental methods used to study the kinetics of proteins searching for target sites on DNA.
Copyright © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures




Similar articles
-
New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.Structure. 2018 Sep 4;26(9):1237-1250.e6. doi: 10.1016/j.str.2018.06.006. Epub 2018 Jul 26. Structure. 2018. PMID: 30057026
-
Sequence-dependent sliding kinetics of p53.Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16552-7. doi: 10.1073/pnas.1120452109. Epub 2012 Sep 25. Proc Natl Acad Sci U S A. 2012. PMID: 23012405 Free PMC article.
-
One-Dimensional Search Dynamics of Tumor Suppressor p53 Regulated by a Disordered C-Terminal Domain.Biophys J. 2017 Jun 6;112(11):2301-2314. doi: 10.1016/j.bpj.2017.04.038. Biophys J. 2017. PMID: 28591603 Free PMC article.
-
Recognition of Local DNA Structures by p53 Protein.Int J Mol Sci. 2017 Feb 10;18(2):375. doi: 10.3390/ijms18020375. Int J Mol Sci. 2017. PMID: 28208646 Free PMC article. Review.
-
The Rich World of p53 DNA Binding Targets: The Role of DNA Structure.Int J Mol Sci. 2019 Nov 9;20(22):5605. doi: 10.3390/ijms20225605. Int J Mol Sci. 2019. PMID: 31717504 Free PMC article. Review.
Cited by
-
Apparent anomalous diffusion and non-Gaussian distributions in a simple mobile-immobile transport model with Poissonian switching.J R Soc Interface. 2022 Jul;19(192):20220233. doi: 10.1098/rsif.2022.0233. Epub 2022 Jul 6. J R Soc Interface. 2022. PMID: 35857918 Free PMC article.
-
Learning the Regulatory Code of Gene Expression.Front Mol Biosci. 2021 Jun 10;8:673363. doi: 10.3389/fmolb.2021.673363. eCollection 2021. Front Mol Biosci. 2021. PMID: 34179082 Free PMC article. Review.
-
Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo.Nucleic Acids Res. 2014 Aug;42(14):8861-72. doi: 10.1093/nar/gku627. Epub 2014 Jul 17. Nucleic Acids Res. 2014. PMID: 25034688 Free PMC article.
-
Speeding up biomolecular interactions by molecular sledding.Chem Sci. 2016 Feb 1;7(2):916-920. doi: 10.1039/C5SC03063C. Epub 2015 Oct 7. Chem Sci. 2016. PMID: 26913169 Free PMC article.
-
Obstacles may facilitate and direct DNA search by proteins.Biophys J. 2013 May 7;104(9):2042-50. doi: 10.1016/j.bpj.2013.03.030. Biophys J. 2013. PMID: 23663847 Free PMC article.
References
-
- Adam G, Delbrück M. In: Reduction of dimensionality in biological difusion processes. Rich A, Davidson N, editors. W.H. Freeman & Company; San Fransisco: 1968. pp. 198–215.
-
- Riggs AD, Bourgeois S, Cohn M. J Mol Biol. 1970;53:401–417. - PubMed
-
- Berg OG, von Hippel PH. Annu Rev Biophys Biophys Chem. 1985;14:131–160. - PubMed
-
- Richter PH, Eigen M. Biophys Chem. 1974;2:255–263. - PubMed
-
- Berg OG, Blomberg C. Biophys Chem. 1976;4:367–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

