Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system
- PMID: 21547297
- PMCID: PMC5938088
- DOI: 10.1039/c1mb05061c
Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system
Abstract
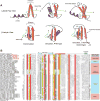
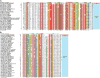
Recent studies point to a diverse assemblage of prokaryotic cognates of the eukaryotic ubiquitin (Ub) system. These systems span an entire spectrum, ranging from those catalyzing cofactor and amino acid biosynthesis, with only adenylating E1-like enzymes and ubiquitin-like proteins (Ubls), to those that are closer to eukaryotic systems by virtue of possessing E2 enzymes. Until recently E3 enzymes were unknown in such prokaryotic systems. Using contextual information from comparative genomics, we uncover a diverse group of RING finger E3s in prokaryotes that are likely to function with E1s, E2s, JAB domain peptidases and Ubls. These E1s, E2s and RING fingers suggest that features hitherto believed to be unique to eukaryotic versions of these proteins emerged progressively in such prokaryotic systems. These include the specific configuration of residues associated with oxyanion-hole formation in E2s and the C-terminal UFD in the E1 enzyme, which presents the E2 to its active site. Our study suggests for the first time that YukD-like Ubls might be conjugated by some of these systems in a manner similar to eukaryotic Ubls. We also show that prokaryotic RING fingers possess considerable functional diversity and that not all of them are involved in Ub-related functions. In eukaryotes, other than RING fingers, a number of distinct binuclear (chelating two Zn atoms) and mononuclear (chelating one zinc atom) treble clef domains are involved in Ub-related functions. Through detailed structural analysis we delineated the higher order relationships and interaction modes of binuclear treble clef domains. This indicated that the FYVE domain acquired the binuclear state independently of the other binuclear forms and that different treble clef domains have convergently acquired Ub-related functions independently of the RING finger. Among these, we uncover evidence for notable prokaryotic radiations of the ZF-UBP, B-box, AN1 and LIM clades of treble clef domains and present contextual evidence to support their role in functions unrelated to the Ub-system in prokaryotes. In particular, we show that bacterial ZF-UBP domains are part of a novel cyclic nucleotide-dependent redox signaling system, whereas prokaryotic B-box, AN1 and LIM domains have related functions as partners of diverse membrane-associated peptidases in processing proteins. This information, in conjunction with structural analysis, suggests that these treble clef domains might have been independently recruited to the eukaryotic Ub-system due to an ancient conserved mode of interaction with peptides.
Figures


Similar articles
-
The UBR-box and its relationship to binuclear RING-like treble clef zinc fingers.Biol Direct. 2015 Jul 17;10:36. doi: 10.1186/s13062-015-0066-5. Biol Direct. 2015. PMID: 26185100 Free PMC article.
-
Structure and evolution of ubiquitin and ubiquitin-related domains.Methods Mol Biol. 2012;832:15-63. doi: 10.1007/978-1-61779-474-2_2. Methods Mol Biol. 2012. PMID: 22350875
-
The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains.Genome Biol. 2006;7(7):R60. doi: 10.1186/gb-2006-7-7-r60. Genome Biol. 2006. PMID: 16859499 Free PMC article.
-
The natural history of ubiquitin and ubiquitin-related domains.Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1433-60. doi: 10.2741/3996. Front Biosci (Landmark Ed). 2012. PMID: 22201813 Free PMC article. Review.
-
NEDD8 and ubiquitin ligation by cullin-RING E3 ligases.Curr Opin Struct Biol. 2021 Apr;67:101-109. doi: 10.1016/j.sbi.2020.10.007. Epub 2020 Nov 5. Curr Opin Struct Biol. 2021. PMID: 33160249 Free PMC article. Review.
Cited by
-
The exportomer: the peroxisomal receptor export machinery.Cell Mol Life Sci. 2013 Apr;70(8):1393-411. doi: 10.1007/s00018-012-1136-9. Epub 2012 Sep 15. Cell Mol Life Sci. 2013. PMID: 22983384 Free PMC article. Review.
-
A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena.Mol Biol Cell. 2017 Mar 15;28(6):825-833. doi: 10.1091/mbc.E16-09-0678. Epub 2017 Jan 18. Mol Biol Cell. 2017. PMID: 28100637 Free PMC article.
-
Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.PLoS One. 2016 Sep 23;11(9):e0163660. doi: 10.1371/journal.pone.0163660. eCollection 2016. PLoS One. 2016. PMID: 27662200 Free PMC article.
-
The UBR-box and its relationship to binuclear RING-like treble clef zinc fingers.Biol Direct. 2015 Jul 17;10:36. doi: 10.1186/s13062-015-0066-5. Biol Direct. 2015. PMID: 26185100 Free PMC article.
-
Diversifying Evolution of the Ubiquitin-26S Proteasome System in Brassicaceae and Poaceae.Int J Mol Sci. 2019 Jun 30;20(13):3226. doi: 10.3390/ijms20133226. Int J Mol Sci. 2019. PMID: 31262075 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

