Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals
- PMID: 21541345
- PMCID: PMC3081832
- DOI: 10.1371/journal.pone.0019040
Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals
Abstract
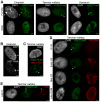
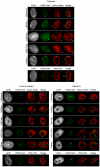
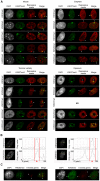
X chromosome inactivation (XCI) is the transcriptional silencing of one X in female mammals, balancing expression of X genes between females (XX) and males (XY). In placental mammals non-coding XIST RNA triggers silencing of one X (Xi) and recruits a characteristic suite of epigenetic modifications, including the histone mark H3K27me3. In marsupials, where XIST is missing, H3K27me3 association seems to have different degrees of stability, depending on cell-types and species. However, the complete suite of histone marks associated with the Xi and their stability throughout cell cycle remain a mystery, as does the evolution of an ancient mammal XCI system. Our extensive immunofluorescence analysis (using antibodies against specific histone modifications) in nuclei of mammals distantly related to human and mouse, revealed a general absence from the mammalian Xi territory of transcription machinery and histone modifications associated with active chromatin. Specific repressive modifications associated with XCI in human and mouse were also observed in elephant (a distantly related placental mammal), as was accumulation of XIST RNA. However, in two marsupial species the Xi either lacked these modifications (H4K20me1), or they were restricted to specific windows of the cell cycle (H3K27me3, H3K9me2). Surprisingly, the marsupial Xi was stably enriched for modifications associated with constitutive heterochromatin in all eukaryotes (H4K20me3, H3K9me3). We propose that marsupial XCI is comparable to a system that evolved in the common therian (marsupial and placental) ancestor. Silent chromatin of the early inactive X was exapted from neighbouring constitutive heterochromatin and, in early placental evolution, was augmented by the rise of XIST and the stable recruitment of specific histone modifications now classically associated with XCI.
Conflict of interest statement
Figures

Similar articles
-
Evolutionary diversity and developmental regulation of X-chromosome inactivation.Hum Genet. 2011 Aug;130(2):307-27. doi: 10.1007/s00439-011-1029-2. Epub 2011 Jun 18. Hum Genet. 2011. PMID: 21687993 Free PMC article. Review.
-
High-resolution analysis of epigenetic changes associated with X inactivation.Genome Res. 2009 Aug;19(8):1361-73. doi: 10.1101/gr.092643.109. Epub 2009 Jul 6. Genome Res. 2009. PMID: 19581487 Free PMC article.
-
Specific patterns of histone marks accompany X chromosome inactivation in a marsupial.Chromosome Res. 2009;17(1):115-26. doi: 10.1007/s10577-009-9020-7. Epub 2009 Feb 13. Chromosome Res. 2009. PMID: 19214764
-
The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients.Proc Natl Acad Sci U S A. 2021 Jun 15;118(24):e2024624118. doi: 10.1073/pnas.2024624118. Proc Natl Acad Sci U S A. 2021. PMID: 34103397 Free PMC article.
-
Xist regulation and function explored.Hum Genet. 2011 Aug;130(2):223-36. doi: 10.1007/s00439-011-1008-7. Epub 2011 May 28. Hum Genet. 2011. PMID: 21626138 Free PMC article. Review.
Cited by
-
Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation.Nature. 2012 Jul 12;487(7406):254-8. doi: 10.1038/nature11171. Nature. 2012. PMID: 22722828 Free PMC article.
-
Imprinted X chromosome inactivation in marsupials: The paternal X arrives at the egg with a silent DNA methylation profile.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2412185121. doi: 10.1073/pnas.2412185121. Epub 2024 Aug 27. Proc Natl Acad Sci U S A. 2024. PMID: 39190362 Free PMC article.
-
Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals.PLoS Genet. 2013;9(7):e1003635. doi: 10.1371/journal.pgen.1003635. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874231 Free PMC article.
-
Paternal X inactivation does not correlate with X chromosome evolutionary strata in marsupials.BMC Evol Biol. 2014 Dec 24;14:267. doi: 10.1186/s12862-014-0267-z. BMC Evol Biol. 2014. PMID: 25539578 Free PMC article.
-
The Methylome of Vertebrate Sex Chromosomes.Genes (Basel). 2018 May 1;9(5):230. doi: 10.3390/genes9050230. Genes (Basel). 2018. PMID: 29723955 Free PMC article. Review.
References
-
- Lyon MF. Gene action in the X-chromosome of the mouse. Nature. 1961;190:372–373. - PubMed
-
- Graves JAM, Gartler SM. Mammalian X chromosome inactivation: testing the hypothesis of transcriptional control. Somat Cell Mol Genet. 1986;12:275–280. - PubMed
-
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. - PubMed
-
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. - PubMed
-
- Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

