Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb)
- PMID: 21536667
- PMCID: PMC3123043
- DOI: 10.1074/jbc.M111.235077
Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb)
Abstract
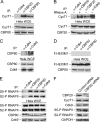
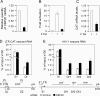
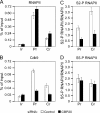
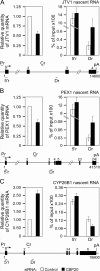
Promoter-proximal pausing of RNAPII coincides with the formation of the cap structure at the 5' end of pre-mRNA, which is bound by the cap-binding protein complex (CBC). Although the positive transcription elongation factor b (P-TEFb) stimulates the release of RNAPII from pausing and promotes transcription elongation and alternative splicing by phosphorylating the RNAPII C-terminal domain at Ser2 (S2-P RNAPII), it is unknown whether CBC facilitates these events. In this study, we report that CBC interacts with P-TEFb and transcriptionally engaged RNAPII and is globally required for optimal levels of S2-P RNAPII. Quantitative nascent RNA immunoprecipitation and ChIP experiments reveal that depletion of CBC attenuates HIV-1 Tat transactivation and impedes transcription elongation of investigated CBC-dependent endogenous genes by decreasing the levels of P-TEFb and S2-P RNAPII, leading to accumulation of RNAPII in the body of these genes. Finally, CBC is essential for the promotion of alternative splicing through facilitating P-TEFb, S2-P RNAPII, and splicing factor 2/alternative splicing factor occupancy at a splicing minigene. These findings disclose a vital role of CBC in connecting pre-mRNA capping to transcription elongation and alternative splicing via P-TEFb.
Figures

Similar articles
-
A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat.Genes Dev. 2005 May 15;19(10):1211-26. doi: 10.1101/gad.1291705. Genes Dev. 2005. PMID: 15905409 Free PMC article.
-
P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms.RNA Biol. 2010 Mar-Apr;7(2):145-50. doi: 10.4161/rna.7.2.11057. Epub 2010 Mar 29. RNA Biol. 2010. PMID: 20305375 Review.
-
7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development.Proc Natl Acad Sci U S A. 2009 May 12;106(19):7798-803. doi: 10.1073/pnas.0903188106. Epub 2009 Apr 27. Proc Natl Acad Sci U S A. 2009. PMID: 19416841 Free PMC article.
-
RBM22 regulates RNA polymerase II 5' pausing, elongation rate, and termination by coordinating 7SK-P-TEFb complex and SPT5.Genome Biol. 2024 Apr 19;25(1):102. doi: 10.1186/s13059-024-03242-6. Genome Biol. 2024. PMID: 38641822 Free PMC article.
-
P-TEFb: The master regulator of transcription elongation.Mol Cell. 2023 Feb 2;83(3):393-403. doi: 10.1016/j.molcel.2022.12.006. Epub 2023 Jan 3. Mol Cell. 2023. PMID: 36599353 Free PMC article. Review.
Cited by
-
A competitive regulatory mechanism discriminates between juxtaposed splice sites and pri-miRNA structures.Nucleic Acids Res. 2013 Oct;41(18):8680-91. doi: 10.1093/nar/gkt614. Epub 2013 Jul 17. Nucleic Acids Res. 2013. PMID: 23863840 Free PMC article.
-
Control of HIV latency by epigenetic and non-epigenetic mechanisms.Curr HIV Res. 2011 Dec 1;9(8):554-67. doi: 10.2174/157016211798998736. Curr HIV Res. 2011. PMID: 22211660 Free PMC article.
-
The Cap-Binding Complex CBC and the Eukaryotic Translation Factor eIF4E: Co-Conspirators in Cap-Dependent RNA Maturation and Translation.Cancers (Basel). 2021 Dec 8;13(24):6185. doi: 10.3390/cancers13246185. Cancers (Basel). 2021. PMID: 34944805 Free PMC article. Review.
-
Ars2 promotes proper replication-dependent histone mRNA 3' end formation.Mol Cell. 2012 Jan 13;45(1):87-98. doi: 10.1016/j.molcel.2011.12.020. Mol Cell. 2012. PMID: 22244333 Free PMC article.
-
Comprehensive investigation in oncogenic functions and immunological roles of NCBP2 and its validation in prostate cancer.Transl Oncol. 2024 Sep;47:102049. doi: 10.1016/j.tranon.2024.102049. Epub 2024 Jul 3. Transl Oncol. 2024. PMID: 38964031 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

