The specific binding to 21-nt double-stranded RNAs is crucial for the anti-silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations
- PMID: 21531919
- PMCID: PMC3096046
- DOI: 10.1261/rna.2510611
The specific binding to 21-nt double-stranded RNAs is crucial for the anti-silencing activity of Cucumber vein yellowing virus P1b and perturbs endogenous small RNA populations
Abstract
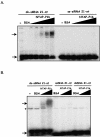
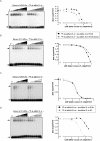
RNA silencing mediated by siRNAs plays an important role as an anti-viral defense mechanism in plants and other eukaryotic organisms, which is usually counteracted by viral RNA silencing suppressors (RSSs). The ipomovirus Cucumber vein yellowing virus (CVYV) lacks the typical RSS of members of the family Potyviridae, HCPro, which is replaced by an unrelated RSS, P1b. CVYV P1b resembles potyviral HCPro in forming complexes with synthetic siRNAs in vitro. Electrophoretic mobility shift assays showed that P1b, like potyviral HCPro, interacts with double-stranded siRNAs, but is not able to bind single-stranded small RNAs or small DNAs. These assays also showed a preference of CVYV P1b for binding to 21-nt siRNAs, a feature also reported for HCPro. However, these two potyvirid RSSs differ in their requirements of 2-nucleotide (nt) 3' overhangs and 5' terminal phosphoryl groups for siRNA binding. Copurification assays confirmed in vivo P1b-siRNA interactions. We have demonstrated by deep sequencing of small RNA populations interacting in vivo with CVYV P1b that the size preference of P1b for small RNAs of 21 nt also takes place in the plant, and that expression of this RSS causes drastic changes in the endogenous small RNA populations. In addition, a site-directed mutagenesis analysis strongly supported the assumption that P1b-siRNA binding is decisive for the anti-silencing activity of P1b and localized a basic domain involved in the siRNA-binding activity of this protein.
Figures





Similar articles
-
Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from cucumber vein yellowing ipomovirus.J Virol. 2008 Jan;82(2):974-86. doi: 10.1128/JVI.01664-07. Epub 2007 Nov 7. J Virol. 2008. PMID: 17989179 Free PMC article.
-
RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro.J Virol. 2006 Oct;80(20):10055-63. doi: 10.1128/JVI.00985-06. J Virol. 2006. PMID: 17005683 Free PMC article.
-
The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host-specific manner.Mol Plant Microbe Interact. 2012 Feb;25(2):151-64. doi: 10.1094/MPMI-08-11-0216. Mol Plant Microbe Interact. 2012. PMID: 21970691
-
The role of the 5' untranslated regions of Potyviridae in translation.Virus Res. 2015 Aug 3;206:74-81. doi: 10.1016/j.virusres.2015.02.005. Epub 2015 Feb 12. Virus Res. 2015. PMID: 25683508 Review.
-
RNA-based viral immunity initiated by the Dicer family of host immune receptors.Immunol Rev. 2009 Jan;227(1):176-88. doi: 10.1111/j.1600-065X.2008.00722.x. Immunol Rev. 2009. PMID: 19120484 Free PMC article. Review.
Cited by
-
The VP3 factor from viruses of Birnaviridae family suppresses RNA silencing by binding both long and small RNA duplexes.PLoS One. 2012;7(9):e45957. doi: 10.1371/journal.pone.0045957. Epub 2012 Sep 25. PLoS One. 2012. PMID: 23049903 Free PMC article.
-
A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles.J Virol. 2014 Sep 1;88(17):9808-18. doi: 10.1128/JVI.01010-14. Epub 2014 Jun 18. J Virol. 2014. PMID: 24942578 Free PMC article.
-
Wheat streak mosaic virus P1 Binds to dsRNAs without Size and Sequence Specificity and a GW Motif Is Crucial for Suppression of RNA Silencing.Viruses. 2019 May 24;11(5):472. doi: 10.3390/v11050472. Viruses. 2019. PMID: 31137615 Free PMC article.
-
Analysis of the RNA-Dependent RNA Polymerase 1 (RDR1) Gene Family in Melon.Plants (Basel). 2022 Jul 7;11(14):1795. doi: 10.3390/plants11141795. Plants (Basel). 2022. PMID: 35890429 Free PMC article.
-
Biochemical and genetic functional dissection of the P38 viral suppressor of RNA silencing.RNA. 2017 May;23(5):639-654. doi: 10.1261/rna.060434.116. Epub 2017 Feb 1. RNA. 2017. PMID: 28148824 Free PMC article.
References
-
- Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 85: 983–991 - PubMed
-
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR 2005. Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat Struct Mol Biol 12: 952–957 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
