Effects of in ovo electroporation on endogenous gene expression: genome-wide analysis
- PMID: 21527010
- PMCID: PMC3105949
- DOI: 10.1186/1749-8104-6-17
Effects of in ovo electroporation on endogenous gene expression: genome-wide analysis
Abstract
Background: In ovo electroporation is a widely used technique to study gene function in developmental biology. Despite the widespread acceptance of this technique, no genome-wide analysis of the effects of in ovo electroporation, principally the current applied across the tissue and exogenous vector DNA introduced, on endogenous gene expression has been undertaken. Here, the effects of electric current and expression of a GFP-containing construct, via electroporation into the midbrain of Hamburger-Hamilton stage 10 chicken embryos, are analysed by microarray.
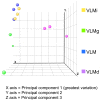
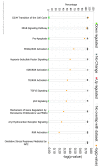
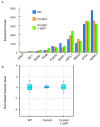
Results: Both current alone and in combination with exogenous DNA expression have a small but reproducible effect on endogenous gene expression, changing the expression of the genes represented on the array by less than 0.1% (current) and less than 0.5% (current + DNA), respectively. The subset of genes regulated by electric current and exogenous DNA span a disparate set of cellular functions. However, no genes involved in the regional identity were affected. In sharp contrast to this, electroporation of a known transcription factor, Dmrt5, caused a much greater change in gene expression.
Conclusions: These findings represent the first systematic genome-wide analysis of the effects of in ovo electroporation on gene expression during embryonic development. The analysis reveals that this process has minimal impact on the genetic basis of cell fate specification. Thus, the study demonstrates the validity of the in ovo electroporation technique to study gene function and expression during development. Furthermore, the data presented here can be used as a resource to refine the set of transcriptional responders in future in ovo electroporation studies of specific gene function.
Figures

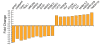

Similar articles
-
Targeted gene expression in the chicken eye by in ovo electroporation.Mol Vis. 2004 Nov 17;10:874-83. Mol Vis. 2004. PMID: 15570216
-
In ovo electroporation in chick midbrain for studying gene function in dopaminergic neuron development.J Vis Exp. 2012 Aug 3;(66):e4017. doi: 10.3791/4017. J Vis Exp. 2012. PMID: 22895156 Free PMC article.
-
In ovo electroporation in embryonic chick retina.J Vis Exp. 2012 Feb 5;(60):3792. doi: 10.3791/3792. J Vis Exp. 2012. PMID: 22330044 Free PMC article.
-
A primer on using in ovo electroporation to analyze gene function.Dev Dyn. 2004 Mar;229(3):433-9. doi: 10.1002/dvdy.10473. Dev Dyn. 2004. PMID: 14991698 Review.
-
'Shocking' developments in chick embryology: electroporation and in ovo gene expression.Nat Cell Biol. 1999 Dec;1(8):E203-7. doi: 10.1038/70231. Nat Cell Biol. 1999. PMID: 10587659 Review.
Cited by
-
Development of retroviral vectors for tissue-restricted expression in chicken embryonic gonads.PLoS One. 2014 Jul 8;9(7):e101811. doi: 10.1371/journal.pone.0101811. eCollection 2014. PLoS One. 2014. PMID: 25003592 Free PMC article.
-
A reliable and flexible gene manipulation strategy in posthatch zebra finch brain.Sci Rep. 2017 Feb 24;7:43244. doi: 10.1038/srep43244. Sci Rep. 2017. PMID: 28233828 Free PMC article.
-
In Ovo Electroporation of Plasmid DNA and Morpholinos into Specific Tissues During Early Embryogenesis.Methods Mol Biol. 2019;1976:71-82. doi: 10.1007/978-1-4939-9412-0_6. Methods Mol Biol. 2019. PMID: 30977066 Free PMC article.
References
-
- Islam SM, Shinmyo Y, Okafuji T, Su Y, Naser IB, Ahmed G, Zhang S, Chen S, Ohta K, Kiyonari H, Abe T, Tanaka S, Nishinakamura R, Terashima T, Kitamura T, Tanaka H. Draxin, a repulsive guidance protein for spinal cord and forebrain commissures. Science. 2009;323:388–393. doi: 10.1126/science.1165187. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

