Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability
- PMID: 21525337
- PMCID: PMC3126497
- DOI: 10.1128/JVI.00246-11
Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability
Abstract
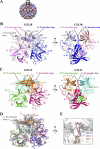
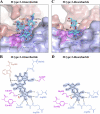
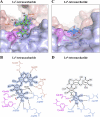
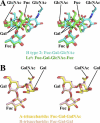
Noroviruses are the dominant cause of outbreaks of gastroenteritis worldwide, and interactions with human histo-blood group antigens (HBGAs) are thought to play a critical role in their entry mechanism. Structures of noroviruses from genogroups GI and GII in complex with HBGAs, however, reveal different modes of interaction. To gain insight into norovirus recognition of HBGAs, we determined crystal structures of norovirus protruding domains from two rarely detected GII genotypes, GII.10 and GII.12, alone and in complex with a panel of HBGAs, and analyzed structure-function implications related to conservation of the HBGA binding pocket. The GII.10- and GII.12-apo structures as well as the previously solved GII.4-apo structure resembled each other more closely than the GI.1-derived structure, and all three GII structures showed similar modes of HBGA recognition. The primary GII norovirus-HBGA interaction involved six hydrogen bonds between a terminal αfucose1-2 of the HBGAs and a dimeric capsid interface, which was composed of elements from two protruding subdomains. Norovirus interactions with other saccharide units of the HBGAs were variable and involved fewer hydrogen bonds. Sequence analysis revealed a site of GII norovirus sequence conservation to reside under the critical αfucose1-2 and to be one of the few patches of conserved residues on the outer virion-capsid surface. The site was smaller than that involved in full HBGA recognition, a consequence of variable recognition of peripheral saccharides. Despite this evasion tactic, the HBGA site of viral vulnerability may provide a viable target for small molecule- and antibody-mediated neutralization of GII norovirus.
Figures


Similar articles
-
Structural basis for norovirus inhibition and fucose mimicry by citrate.J Virol. 2012 Jan;86(1):284-92. doi: 10.1128/JVI.05909-11. Epub 2011 Oct 26. J Virol. 2012. PMID: 22031945 Free PMC article.
-
Human noroviruses' fondness for histo-blood group antigens.J Virol. 2015 Feb;89(4):2024-40. doi: 10.1128/JVI.02968-14. Epub 2014 Nov 26. J Virol. 2015. PMID: 25428879 Free PMC article.
-
Epitope mapping of histo blood group antigens bound to norovirus VLPs using STD NMR experiments reveals fine details of molecular recognition.Glycoconj J. 2017 Oct;34(5):679-689. doi: 10.1007/s10719-017-9792-5. Epub 2017 Aug 19. Glycoconj J. 2017. PMID: 28823097
-
[Noroviruses--tactic of spread].Przegl Epidemiol. 2009;63(1):5-9. Przegl Epidemiol. 2009. PMID: 19522218 Review. Polish.
-
Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations.Immunol Rev. 2008 Oct;225:190-211. doi: 10.1111/j.1600-065X.2008.00680.x. Immunol Rev. 2008. PMID: 18837783 Review.
Cited by
-
Structural basis for norovirus neutralization by an HBGA blocking human IgA antibody.Proc Natl Acad Sci U S A. 2016 Oct 4;113(40):E5830-E5837. doi: 10.1073/pnas.1609990113. Epub 2016 Sep 19. Proc Natl Acad Sci U S A. 2016. PMID: 27647885 Free PMC article.
-
Structural Constraints on Human Norovirus Binding to Histo-Blood Group Antigens.mSphere. 2016 Mar 30;1(2):e00049-16. doi: 10.1128/mSphere.00049-16. eCollection 2016 Mar-Apr. mSphere. 2016. PMID: 27303720 Free PMC article.
-
First detection of neboviruses in yak (Bos grunniens) and identification of a novel neboviruses based on complete genome.Vet Microbiol. 2019 Sep;236:108388. doi: 10.1016/j.vetmic.2019.108388. Epub 2019 Aug 9. Vet Microbiol. 2019. PMID: 31500726 Free PMC article.
-
Identification and characterization of antibody-binding epitopes on the norovirus GII.3 capsid.J Virol. 2014 Feb;88(4):1942-52. doi: 10.1128/JVI.02992-13. Epub 2013 Nov 27. J Virol. 2014. PMID: 24284328 Free PMC article.
-
Structural basis of glycan interaction in gastroenteric viral pathogens.Curr Opin Virol. 2014 Aug;7:119-27. doi: 10.1016/j.coviro.2014.05.008. Epub 2014 Jul 27. Curr Opin Virol. 2014. PMID: 25073118 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

