Subversion of the actin cytoskeleton during viral infection
- PMID: 21522191
- PMCID: PMC3229036
- DOI: 10.1038/nrmicro2574
Subversion of the actin cytoskeleton during viral infection
Abstract
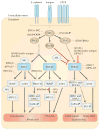
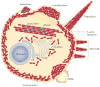
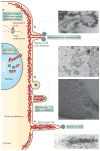
Viral infection converts the normal functions of a cell to optimize viral replication and virion production. One striking observation of this conversion is the reconfiguration and reorganization of cellular actin, affecting every stage of the viral life cycle, from entry through assembly to egress. The extent and degree of cytoskeletal reorganization varies among different viral infections, suggesting the evolution of myriad viral strategies. In this Review, we describe how the interaction of viral proteins with the cell modulates the structure and function of the actin cytoskeleton to initiate, sustain and spread infections. The molecular biology of such interactions continues to engage virologists in their quest to understand viral replication and informs cell biologists about the role of the cytoskeleton in the uninfected cell.
Figures


Similar articles
-
Imaging, Tracking and Computational Analyses of Virus Entry and Egress with the Cytoskeleton.Viruses. 2018 Mar 31;10(4):166. doi: 10.3390/v10040166. Viruses. 2018. PMID: 29614729 Free PMC article. Review.
-
Actin and Rho GTPases in herpesvirus biology.Trends Microbiol. 2007 Sep;15(9):426-33. doi: 10.1016/j.tim.2007.08.003. Epub 2007 Aug 30. Trends Microbiol. 2007. PMID: 17764949 Review.
-
Actin' up: herpesvirus interactions with Rho GTPase signaling.Viruses. 2011 Apr;3(4):278-92. doi: 10.3390/v3040278. Epub 2011 Mar 24. Viruses. 2011. PMID: 21994732 Free PMC article. Review.
-
Differential effects of actin cytoskeleton dynamics on equine infectious anemia virus particle production.J Virol. 2004 Jan;78(2):882-91. doi: 10.1128/jvi.78.2.882-891.2004. J Virol. 2004. PMID: 14694119 Free PMC article.
-
Actin in herpesvirus infection.Viruses. 2011 Apr;3(4):336-46. doi: 10.3390/v3040336. Epub 2011 Apr 12. Viruses. 2011. PMID: 21994736 Free PMC article. Review.
Cited by
-
High throughput proteomic analysis and a comparative review identify the nuclear chaperone, Nucleophosmin among the common set of proteins modulated in Chikungunya virus infection.J Proteomics. 2015 Apr 29;120:126-41. doi: 10.1016/j.jprot.2015.03.007. Epub 2015 Mar 14. J Proteomics. 2015. PMID: 25782748 Free PMC article. Review.
-
Mechanisms of budding of nanoscale particles through lipid bilayers.J Phys Chem B. 2012 Aug 16;116(32):9595-603. doi: 10.1021/jp301601g. Epub 2012 Aug 3. J Phys Chem B. 2012. PMID: 22803595 Free PMC article.
-
The actin cytoskeleton is important for rotavirus internalization and RNA genome replication.Virus Res. 2019 Apr 2;263:27-33. doi: 10.1016/j.virusres.2019.01.003. Epub 2019 Jan 9. Virus Res. 2019. PMID: 30639190 Free PMC article.
-
Arp2/3-mediated actin-based motility: a tail of pathogen abuse.Cell Host Microbe. 2013 Sep 11;14(3):242-55. doi: 10.1016/j.chom.2013.08.011. Cell Host Microbe. 2013. PMID: 24034611 Free PMC article. Review.
-
6-Thioguanine Inhibits Herpes Simplex Virus 1 Infection of Eyes.Microbiol Spectr. 2021 Dec 22;9(3):e0064621. doi: 10.1128/Spectrum.00646-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730435 Free PMC article.
References
-
- Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. - PubMed
-
- Moreau V, et al. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nature Cell Biol. 2000;2:441–448. - PubMed
-
- Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

