Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3
- PMID: 21498885
- PMCID: PMC3122172
- DOI: 10.1074/jbc.M111.237636
Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3
Abstract
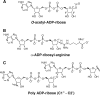
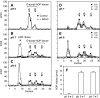
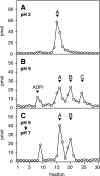
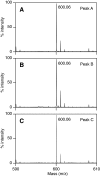
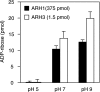
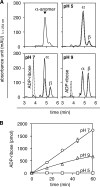
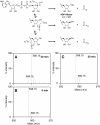
O-acetyl-ADP-ribose (OAADPr), produced by the Sir2-catalyzed NAD(+)-dependent histone/protein deacetylase reaction, regulates diverse biological processes. Interconversion between two OAADPr isomers with acetyl attached to the C-2″ and C-3″ hydroxyl of ADP-ribose (ADPr) is rapid. We reported earlier that ADP-ribosylhydrolase 3 (ARH3), one of three ARH proteins sharing structural similarities, hydrolyzed OAADPr to ADPr and acetate, and poly(ADPr) to ADPr monomers. ARH1 also hydrolyzed OAADPr and poly(ADPr) as well as ADP-ribose-arginine, with arginine in α-anomeric linkage to C-1″ of ADP-ribose. Because both ARH3- and ARH1-catalyzed reactions involve nucleophilic attacks at the C-1″ position, it was perplexing that the ARH3 catalytic site would cleave OAADPr at either the 2″- or 3″-position, and we postulated the existence of a third isomer, 1″-OAADPr, in equilibrium with 2″- and 3″-isomers. A third isomer, consistent with 1″-OAADPr, was identified at pH 9.0. Further, ARH3 OAADPr hydrolase activity was greater at pH 9.0 than at neutral pH where 3″-OAADPr predominated. Consistent with our hypothesis, IC(50) values for ARH3 inhibition by 2″- and 3″-N-acetyl-ADPr analogs of OAADPr were significantly higher than that for ADPr. ARH1 also hydrolyzed OAADPr more rapidly at alkaline pH, but cleavage of ADP-ribose-arginine was faster at neutral pH than pH 9.0. ARH3-catalyzed hydrolysis of OAADPr in H(2)(18)O resulted in incorporation of one (18)O into ADP-ribose by mass spectrometric analysis, consistent with cleavage at the C-1″ position. Together, these data suggest that ARH family members, ARH1 and ARH3, catalyze hydrolysis of the 1″-O linkage in their structurally diverse substrates.
Figures

Similar articles
-
ARH Family of ADP-Ribose-Acceptor Hydrolases.Cells. 2022 Nov 30;11(23):3853. doi: 10.3390/cells11233853. Cells. 2022. PMID: 36497109 Free PMC article. Review.
-
ADP-Ribosyl-Acceptor Hydrolase Activities Catalyzed by the ARH Family of Proteins.Methods Mol Biol. 2018;1813:187-204. doi: 10.1007/978-1-4939-8588-3_12. Methods Mol Biol. 2018. PMID: 30097868
-
The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases.Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16687-91. doi: 10.1073/pnas.0607911103. Epub 2006 Oct 30. Proc Natl Acad Sci U S A. 2006. PMID: 17075046 Free PMC article.
-
Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose.Biochim Biophys Acta. 2010 Aug;1804(8):1617-25. doi: 10.1016/j.bbapap.2010.02.007. Epub 2010 Feb 20. Biochim Biophys Acta. 2010. PMID: 20176146 Free PMC article. Review.
-
Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases.DNA Repair (Amst). 2014 Nov;23:88-94. doi: 10.1016/j.dnarep.2014.03.005. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746921 Free PMC article. Review.
Cited by
-
The chemistry of the vitamin B3 metabolome.Biochem Soc Trans. 2019 Feb 28;47(1):131-147. doi: 10.1042/BST20180420. Epub 2018 Dec 17. Biochem Soc Trans. 2019. PMID: 30559273 Free PMC article. Review.
-
Functional roles of ADP-ribosylation writers, readers and erasers.Front Cell Dev Biol. 2022 Aug 11;10:941356. doi: 10.3389/fcell.2022.941356. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36035988 Free PMC article. Review.
-
(ADP-ribosyl)hydrolases: structure, function, and biology.Genes Dev. 2020 Mar 1;34(5-6):263-284. doi: 10.1101/gad.334631.119. Epub 2020 Feb 6. Genes Dev. 2020. PMID: 32029451 Free PMC article. Review.
-
Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.Nat Commun. 2021 Jul 28;12(1):4581. doi: 10.1038/s41467-021-24723-3. Nat Commun. 2021. PMID: 34321462 Free PMC article.
-
ARH Family of ADP-Ribose-Acceptor Hydrolases.Cells. 2022 Nov 30;11(23):3853. doi: 10.3390/cells11233853. Cells. 2022. PMID: 36497109 Free PMC article. Review.
References
-
- Williamson K. C., Moss J. (1990) in ADP-ribosylating Toxins and G Proteins: Insights into Signal Transduction (Moss J., Vaughan M. eds) pp. 493–510, American Society for Microbiology, Washington, D.C - PubMed
-
- Fishman P. H. (1990) in ADP-ribosylating Toxins and G Proteins: Insights into Signal Transduction (Moss J., Vaughan M. eds) pp. 127–140, American Society for Microbiology, Washington, D.C - PubMed
-
- Moss J., Zolkiewska A., Okazaki I. (1997) Adv. Exp. Med. Biol. 419, 25–33 - PubMed
-
- Moss J., Tsai S. C., Adamik R., Chen H. C., Stanley S. J. (1988) Biochemistry 27, 5819–5823 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

