Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless
- PMID: 21497759
- PMCID: PMC3087180
- DOI: 10.1016/j.devcel.2011.03.017
Establishment of medial fates along the proximodistal axis of the Drosophila leg through direct activation of dachshund by Distalless
Abstract
The proximodistal (PD) axis of the Drosophila leg is thought to be established by the combined gradients of two secreted morphogens, Wingless (Wg) and Decapentaplegic (Dpp). According to this model, high [Wg+Dpp] activates Distalless (Dll) and represses dachshund (dac) in the distal cells of the leg disc, while intermediate [Wg+Dpp] activates dac in medial tissue. To test this model we identified and characterized a dac cis-regulatory element (dac RE) that recapitulates dac's medial expression domain during leg development. Counter to the gradient model, we find that Wg and Dpp do not act in a graded manner to activate RE. Instead, dac RE is activated directly by Dll and repressed distally by a combination of factors, including the homeodomain protein Bar. Thus, medial leg fates are established via a regulatory cascade in which Wg+Dpp activate Dll and then Dll directly activates dac, with Wg+Dpp as less critical, permissive inputs.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
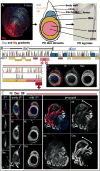
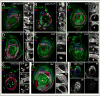
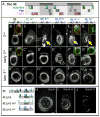
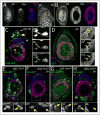
Similar articles
-
Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg.Development. 2008 Feb;135(4):627-36. doi: 10.1242/dev.014670. Epub 2008 Jan 9. Development. 2008. PMID: 18184724 Free PMC article.
-
Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development.Dev Cell. 2008 Jan;14(1):86-96. doi: 10.1016/j.devcel.2007.11.002. Dev Cell. 2008. PMID: 18194655 Free PMC article.
-
A dynamic network of morphogens and transcription factors patterns the fly leg.Curr Top Dev Biol. 2012;98:173-98. doi: 10.1016/B978-0-12-386499-4.00007-0. Curr Top Dev Biol. 2012. PMID: 22305163 Free PMC article. Review.
-
Leg development in flies versus grasshoppers: differences in dpp expression do not lead to differences in the expression of downstream components of the leg patterning pathway.Development. 2000 Apr;127(8):1617-26. doi: 10.1242/dev.127.8.1617. Development. 2000. PMID: 10725238
-
T-box genes organize the dorsal ventral leg axis in Drosophila melanogaster.Fly (Austin). 2010 Apr-Jun;4(2):159-62. doi: 10.4161/fly.4.2.11251. Epub 2010 Apr 18. Fly (Austin). 2010. PMID: 20215860 Review.
Cited by
-
Walking strides direct rapid and flexible recruitment of visual circuits for course control in Drosophila.Neuron. 2022 Jul 6;110(13):2124-2138.e8. doi: 10.1016/j.neuron.2022.04.008. Epub 2022 May 6. Neuron. 2022. PMID: 35525243 Free PMC article.
-
Role of the Forkhead Transcription Factors Fd4 and Fd5 During Drosophila Leg Development.Front Cell Dev Biol. 2021 Aug 2;9:723927. doi: 10.3389/fcell.2021.723927. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34409041 Free PMC article.
-
Early patterning followed by tissue growth establishes distal identity in Drosophila Malpighian tubules.Front Cell Dev Biol. 2022 Aug 19;10:947376. doi: 10.3389/fcell.2022.947376. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36060795 Free PMC article.
-
A faithful internal representation of walking movements in the Drosophila visual system.Nat Neurosci. 2017 Jan;20(1):72-81. doi: 10.1038/nn.4435. Epub 2016 Oct 31. Nat Neurosci. 2017. PMID: 27798632
-
Sp1 modifies leg-to-wing transdetermination in Drosophila.Dev Biol. 2013 Jan 15;373(2):290-9. doi: 10.1016/j.ydbio.2012.11.008. Epub 2012 Nov 17. Dev Biol. 2013. PMID: 23165292 Free PMC article.
References
-
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. - PubMed
-
- Ben-Tabou de-Leon S, Davidson EH. Deciphering the underlying mechanism of specification and differentiation: the sea urchin gene regulatory network. Sci STKE. 2006:pe47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

