During autophagy mitochondria elongate, are spared from degradation and sustain cell viability
- PMID: 21478857
- PMCID: PMC3088644
- DOI: 10.1038/ncb2220
During autophagy mitochondria elongate, are spared from degradation and sustain cell viability
Abstract
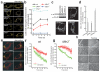
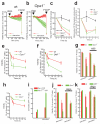
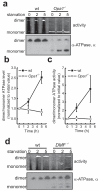
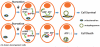
A plethora of cellular processes, including apoptosis, depend on regulated changes in mitochondrial shape and ultrastructure. The role of mitochondria and of their morphology during autophagy, a bulk degradation and recycling process of eukaryotic cells' constituents, is not well understood. Here we show that mitochondrial morphology determines the cellular response to macroautophagy. When autophagy is triggered, mitochondria elongate in vitro and in vivo. During starvation, cellular cyclic AMP levels increase and protein kinase A (PKA) is activated. PKA in turn phosphorylates the pro-fission dynamin-related protein 1 (DRP1), which is therefore retained in the cytoplasm, leading to unopposed mitochondrial fusion. Elongated mitochondria are spared from autophagic degradation, possess more cristae, increased levels of dimerization and activity of ATP synthase, and maintain ATP production. Conversely, when elongation is genetically or pharmacologically blocked, mitochondria consume ATP, precipitating starvation-induced death. Thus, regulated changes in mitochondrial morphology determine the fate of the cell during autophagy.
Figures




Comment in
-
Autophagy: shaping the fate of mitochondria.Nat Rev Mol Cell Biol. 2011 Jun;12(6):344-5. doi: 10.1038/nrm3116. Epub 2011 Apr 29. Nat Rev Mol Cell Biol. 2011. PMID: 21527952 No abstract available.
-
Mitochondria unite to survive.Nat Cell Biol. 2011 May;13(5):521-2. doi: 10.1038/ncb0511-521. Nat Cell Biol. 2011. PMID: 21540850
Similar articles
-
Mitochondrial elongation during autophagy: a stereotypical response to survive in difficult times.Autophagy. 2011 Oct;7(10):1251-3. doi: 10.4161/auto.7.10.16771. Epub 2011 Oct 1. Autophagy. 2011. PMID: 21743300 Free PMC article.
-
Essential amino acids and glutamine regulate induction of mitochondrial elongation during autophagy.Cell Cycle. 2011 Aug 15;10(16):2635-9. doi: 10.4161/cc.10.16.17002. Epub 2011 Aug 15. Cell Cycle. 2011. PMID: 21811092
-
Mitochondria unite to survive.Nat Cell Biol. 2011 May;13(5):521-2. doi: 10.1038/ncb0511-521. Nat Cell Biol. 2011. PMID: 21540850
-
Mitochondrial morphology in mitophagy and macroautophagy.Biochim Biophys Acta. 2013 Jan;1833(1):205-12. doi: 10.1016/j.bbamcr.2012.02.012. Epub 2012 Mar 1. Biochim Biophys Acta. 2013. PMID: 22406072 Review.
-
Regulation of Mitochondrial Dynamics and Autophagy by the Mitochondria-Associated Membrane.Adv Exp Med Biol. 2017;997:33-47. doi: 10.1007/978-981-10-4567-7_3. Adv Exp Med Biol. 2017. PMID: 28815520 Review.
Cited by
-
Cytoprotection by a naturally occurring variant of ATP5G1 in Arctic ground squirrel neural progenitor cells.Elife. 2020 Oct 14;9:e55578. doi: 10.7554/eLife.55578. Elife. 2020. PMID: 33050999 Free PMC article.
-
DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis.J Cell Biol. 2012 May 14;197(4):487-97. doi: 10.1083/jcb.201110058. J Cell Biol. 2012. PMID: 22584906 Free PMC article.
-
Analysis of beta-cell maturity and mitochondrial morphology in juvenile non-human primates exposed to maternal Western-style diet during development.Front Endocrinol (Lausanne). 2024 Jul 24;15:1417437. doi: 10.3389/fendo.2024.1417437. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39114287 Free PMC article.
-
Photobiomodulation in 3D tissue engineering.J Biomed Opt. 2022 Sep;27(9):090901. doi: 10.1117/1.JBO.27.9.090901. J Biomed Opt. 2022. PMID: 36104833 Free PMC article.
-
Impact of C24:0 on actin-microtubule interaction in human neuronal SK-N-BE cells: evaluation by FRET confocal spectral imaging microscopy after dual staining with rhodamine-phalloidin and tubulin tracker green.Funct Neurol. 2015 Jan-Mar;30(1):33-46. Funct Neurol. 2015. PMID: 26214025 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

