Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites
- PMID: 21435340
- PMCID: PMC3273848
- DOI: 10.1016/j.ydbio.2011.03.007
Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites
Abstract
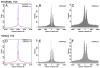
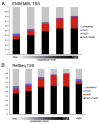
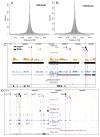
An organism's genome sequence serves as a blueprint for the proteins and regulatory RNAs essential for cellular function. The genome also harbors cis-acting non-coding sequences that control gene expression and are essential to coordinate regulatory programs during embryonic development. However, the genome sequence is largely identical between cell types within a multi-cellular organism indicating that factors such as DNA accessibility and chromatin structure play a crucial role in governing cell-specific gene expression. Recent studies have identified particular chromatin modifications that define functionally distinct cis regulatory elements. Among these are forms of histone 3 that are mono- or tri-methylated at lysine 4 (H3K4me1 or H3K4me3, respectively), which bind preferentially to promoter and enhancer elements in the mammalian genome. In this work, we investigated whether these modified histones could similarly identify cis regulatory elements within the zebrafish genome. By applying chromatin immunoprecipitation followed by deep sequencing, we find that H3K4me1 and H3K4me3 are enriched at transcriptional start sites in the genome of the developing zebrafish embryo and that this association correlates with gene expression. We further find that these modifications associate with distal non-coding conserved elements, including known active enhancers. Finally, we demonstrate that it is possible to utilize H3K4me1 and H3K4me3 binding profiles in combination with available expression data to computationally identify relevant cis regulatory sequences flanking syn-expressed genes in the developing embryo. Taken together, our results indicate that H3K4me1 and H3K4me3 generally mark cis regulatory elements within the zebrafish genome and indicate that further characterization of the zebrafish using this approach will prove valuable in defining transcriptional networks in this model system.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos.Int J Dev Biol. 2010;54(5):803-13. doi: 10.1387/ijdb.103081ll. Int J Dev Biol. 2010. PMID: 20336603
-
Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis.Dev Biol. 2010 Jan 15;337(2):496-505. doi: 10.1016/j.ydbio.2009.10.028. Epub 2009 Nov 3. Dev Biol. 2010. PMID: 19895802
-
A map of cis-regulatory elements and 3D genome structures in zebrafish.Nature. 2020 Dec;588(7837):337-343. doi: 10.1038/s41586-020-2962-9. Epub 2020 Nov 25. Nature. 2020. PMID: 33239788 Free PMC article.
-
Zebrafish embryogenesis - A framework to study regulatory RNA elements in development and disease.Dev Biol. 2020 Jan 15;457(2):172-180. doi: 10.1016/j.ydbio.2019.01.008. Epub 2019 Jan 16. Dev Biol. 2020. PMID: 30659794 Review.
-
Enhancer biology and enhanceropathies.Nat Struct Mol Biol. 2014 Mar;21(3):210-9. doi: 10.1038/nsmb.2784. Nat Struct Mol Biol. 2014. PMID: 24599251 Review.
Cited by
-
Computational methods to detect conserved non-genic elements in phylogenetically isolated genomes: application to zebrafish.Nucleic Acids Res. 2013 Aug;41(15):e151. doi: 10.1093/nar/gkt557. Epub 2013 Jun 27. Nucleic Acids Res. 2013. PMID: 23814184 Free PMC article.
-
Functionally conserved enhancers with divergent sequences in distant vertebrates.BMC Genomics. 2015 Oct 30;16:882. doi: 10.1186/s12864-015-2070-7. BMC Genomics. 2015. PMID: 26519295 Free PMC article.
-
Genetic Dissection of Dual Roles for the Transcription Factor six7 in Photoreceptor Development and Patterning in Zebrafish.PLoS Genet. 2016 Apr 8;12(4):e1005968. doi: 10.1371/journal.pgen.1005968. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27058886 Free PMC article.
-
Memory Macrophages.Int J Mol Sci. 2022 Dec 20;24(1):38. doi: 10.3390/ijms24010038. Int J Mol Sci. 2022. PMID: 36613481 Free PMC article. Review.
-
An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement.Genome Res. 2012 Apr;22(4):642-55. doi: 10.1101/gr.132233.111. Epub 2012 Jan 10. Genome Res. 2012. PMID: 22234889 Free PMC article.
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
-
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

