Genome-virome interactions: examining the role of common viral infections in complex disease
- PMID: 21407242
- PMCID: PMC3678363
- DOI: 10.1038/nrmicro2541
Genome-virome interactions: examining the role of common viral infections in complex disease
Abstract
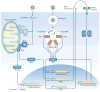
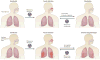
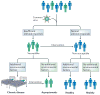
New technologies have widened our view of 'complex diseases': those with both genetic and environmental risk factors. In this Review, we explore recent genetic and virological evidence implicating host-virus interactions in three diseases: type 1 diabetes, inflammatory bowel disease and asthma. The viruses implicated in these diseases cause mucosal infections that affect most of the population but are asymptomatic or mild in many hosts. These findings place a new emphasis on common viral infections as important environmental factors in the pathogenesis of complex diseases, and they compel the field to pursue a better understanding of host interactions with the human virome.
Figures

Similar articles
-
The Role of Viruses in the Pathogenesis of Immune-Mediated Gastro-Intestinal Diseases.Int J Mol Sci. 2024 Jul 30;25(15):8301. doi: 10.3390/ijms25158301. Int J Mol Sci. 2024. PMID: 39125870 Free PMC article. Review.
-
The virome in host health and disease.Immunity. 2015 May 19;42(5):805-13. doi: 10.1016/j.immuni.2015.05.003. Immunity. 2015. PMID: 25992857 Free PMC article. Review.
-
The science of the host-virus network.Nat Microbiol. 2021 Dec;6(12):1483-1492. doi: 10.1038/s41564-021-00999-5. Epub 2021 Nov 24. Nat Microbiol. 2021. PMID: 34819645 Review.
-
Viral manipulation of the host immune response.Curr Opin Immunol. 2015 Oct;36:54-60. doi: 10.1016/j.coi.2015.06.012. Epub 2015 Jul 10. Curr Opin Immunol. 2015. PMID: 26177523 Free PMC article. Review.
-
Chemical Methods for Probing Virus-Host Proteomic Interactions.ACS Infect Dis. 2016 Nov 11;2(11):773-786. doi: 10.1021/acsinfecdis.6b00084. Epub 2016 Jun 29. ACS Infect Dis. 2016. PMID: 27933785 Review.
Cited by
-
Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System.Viruses. 2018 Dec 25;11(1):10. doi: 10.3390/v11010010. Viruses. 2018. PMID: 30585199 Free PMC article. Review.
-
Beyond Cytomegalovirus and Epstein-Barr Virus: a Review of Viruses Composing the Blood Virome of Solid Organ Transplant and Hematopoietic Stem Cell Transplant Recipients.Clin Microbiol Rev. 2020 Aug 26;33(4):e00027-20. doi: 10.1128/CMR.00027-20. Print 2020 Sep 16. Clin Microbiol Rev. 2020. PMID: 32847820 Free PMC article. Review.
-
Beyond research: a primer for considerations on using viral metagenomics in the field and clinic.Front Microbiol. 2015 Mar 25;6:224. doi: 10.3389/fmicb.2015.00224. eCollection 2015. Front Microbiol. 2015. PMID: 25859244 Free PMC article.
-
A Role for the Intestinal Microbiota and Virome in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)?J Clin Med. 2016 Jun 6;5(6):55. doi: 10.3390/jcm5060055. J Clin Med. 2016. PMID: 27275835 Free PMC article. Review.
-
Invasions of Host-Associated Microbiome Networks.Adv Ecol Res. 2017;57:201-281. doi: 10.1016/bs.aecr.2016.11.002. Adv Ecol Res. 2017. PMID: 39404686 Free PMC article.
References
-
- Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Meth Enzymol. 1987;155:335–350. - PubMed
-
- Mackay IM, Mackay JF, Nissen MD, Sloots TP. In: Real-time PCR in Microbiology: From Diagnosis to Characterisation. Mackay IM, editor. Caister Academic Press; Norfolk, U. K: 2007. pp. 1–40.
-
- Monto AS, Cavallaro JJ. The Tecumseh study of respiratory illness. II Patterns of occurrence of infection with respiratory pathogens, 1965–1969. Am J Epidemiol. 1971;94:280–289. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

