Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression
- PMID: 21402699
- PMCID: PMC3089562
- DOI: 10.1074/jbc.M110.205146
Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression
Abstract
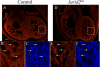
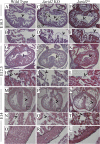
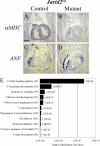
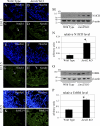
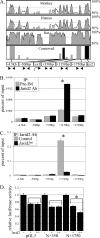
Jarid2/Jumonji critically regulates developmental processes including cardiovascular development. Jarid2 knock-out mice exhibit cardiac defects including hypertrabeculation with noncompaction of the ventricular wall. However, molecular mechanisms underlying Jarid2-mediated cardiac development remain unknown. To determine the cardiac lineage-specific roles of Jarid2, we generated myocardial, epicardial, cardiac neural crest, or endothelial conditional Jarid2 knock-out mice using Cre-loxP technology. Only mice with an endothelial deletion of Jarid2 recapitulate phenotypic defects observed in whole body mutants including hypertrabeculation and noncompaction of the ventricle. To identify potential targets of Jarid2, combinatorial approaches using microarray and candidate gene analyses were employed on Jarid2 knock-out embryonic hearts. Whole body or endothelial deletion of Jarid2 leads to increased endocardial Notch1 expression in the developing ventricle, resulting in increased Notch1-dependent signaling to the adjacent myocardium. Using quantitative chromatin immunoprecipitation analysis, Jarid2 was found to occupy a specific region on the endogenous Notch1 locus. We propose that failure to properly regulate Notch signaling in Jarid2 mutants likely leads to the defects in the developing ventricular chamber. The identification of Jarid2 as a potential regulator of Notch1 signaling has broad implications for many cellular processes including development, stem cell maintenance, and tumor formation.
Figures

Similar articles
-
Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart.J Biol Chem. 2012 Jan 6;287(2):1235-41. doi: 10.1074/jbc.M111.315945. Epub 2011 Nov 21. J Biol Chem. 2012. PMID: 22110129 Free PMC article.
-
Cardiac-specific developmental and epigenetic functions of Jarid2 during embryonic development.J Biol Chem. 2018 Jul 27;293(30):11659-11673. doi: 10.1074/jbc.RA118.002482. Epub 2018 Jun 11. J Biol Chem. 2018. PMID: 29891551 Free PMC article.
-
Myocardial-specific ablation of Jumonji and AT-rich interaction domain-containing 2 (Jarid2) leads to dilated cardiomyopathy in mice.J Biol Chem. 2019 Mar 29;294(13):4981-4996. doi: 10.1074/jbc.RA118.005634. Epub 2019 Jan 30. J Biol Chem. 2019. PMID: 30700554 Free PMC article.
-
Notch signaling as an important mediator of cardiac repair and regeneration after myocardial infarction.Trends Cardiovasc Med. 2010 Oct;20(7):228-31. doi: 10.1016/j.tcm.2011.11.006. Trends Cardiovasc Med. 2010. PMID: 22293023 Free PMC article. Review.
-
Polycomb repressive complex 2 in embryonic stem cells: an overview.Protein Cell. 2010 Dec;1(12):1056-62. doi: 10.1007/s13238-010-0142-7. Epub 2011 Jan 8. Protein Cell. 2010. PMID: 21213100 Free PMC article. Review.
Cited by
-
Epigenetics and Mechanobiology in Heart Development and Congenital Heart Disease.Diseases. 2019 Sep 1;7(3):52. doi: 10.3390/diseases7030052. Diseases. 2019. PMID: 31480510 Free PMC article. Review.
-
Epigenetics.Adv Exp Med Biol. 2024;1441:341-364. doi: 10.1007/978-3-031-44087-8_18. Adv Exp Med Biol. 2024. PMID: 38884720 Review.
-
Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease.JCI Insight. 2017 Oct 19;2(20):e95085. doi: 10.1172/jci.insight.95085. JCI Insight. 2017. PMID: 29046480 Free PMC article.
-
Jarid2 Coordinates Nanog Expression and PCP/Wnt Signaling Required for Efficient ESC Differentiation and Early Embryo Development.Cell Rep. 2015 Jul 28;12(4):573-86. doi: 10.1016/j.celrep.2015.06.060. Epub 2015 Jul 16. Cell Rep. 2015. PMID: 26190104 Free PMC article.
-
Etv2-miR-130a-Jarid2 cascade regulates vascular patterning during embryogenesis.PLoS One. 2017 Dec 12;12(12):e0189010. doi: 10.1371/journal.pone.0189010. eCollection 2017. PLoS One. 2017. PMID: 29232705 Free PMC article.
References
-
- Olson E. N. (2004) Nat. Med. 10, 467–474 - PubMed
-
- Lee Y., Song A. J., Baker R., Micales B., Conway S. J., Lyons G. E. (2000) Circ. Res. 86, 932–938 - PubMed
-
- Jung J., Kim T. G., Lyons G. E., Kim H. R., Lee Y. (2005) J. Biol. Chem. 280, 30916–30923 - PubMed
-
- Balciunas D., Ronne H. (2000) Trends Biochem. Sci. 25, 274–276 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

