STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor
- PMID: 21379341
- PMCID: PMC3040673
- DOI: 10.1371/journal.ppat.1001297
STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor
Abstract
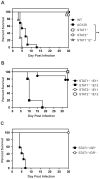
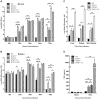
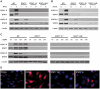
Dengue virus (DENV) is a mosquito-borne flavivirus, and symptoms of infection range from asymptomatic to the severe dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS). High viral loads correlate with disease severity, and both type I & II interferons (IFNs) are crucial for controlling viral replication. We have previously reported that signal transducer and activator of transcription (STAT) 1-deficient mice are resistant to DENV-induced disease, but little is known about this STAT1-independent mechanism of protection. To determine the molecular basis of the STAT1-independent pathway, mice lacking STAT1, STAT2, or both STAT1 and STAT2 were infected with a virulent mouse-adapted strain of DENV2. In the first 72 hours of infection, the single-deficient mice lacking STAT1 or STAT2 possessed 50-100 fold higher levels of viral RNA than wild type mice in the serum, spleen, and other visceral tissues, but remained resistant to DENV-induced death. In contrast, the double-deficient mice exhibited the early death phenotype previously observed in type I and II IFN receptor knockout mice (AG129), indicating that STAT2 is the mediator of the STAT1-independent host defense mechanism. Further studies demonstrated that this STAT2-dependent STAT1-independent mechanism requires the type I IFN receptor, and contributes to the autocrine amplification of type I IFN expression. Examination of gene expression in the spleen and bone marrow-derived macrophages following DENV infection revealed STAT2-dependent pathways can induce the transcription of a subset of interferon stimulated genes even in the absence of STAT1. Collectively, these results help elucidate the nature of the poorly understood STAT1-independent host defense mechanism against viruses by identifying a functional type I IFN/STAT2 signaling pathway following DENV infection in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
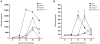

Similar articles
-
The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus.J Immunol. 2013 Oct 15;191(8):4194-201. doi: 10.4049/jimmunol.1300799. Epub 2013 Sep 16. J Immunol. 2013. PMID: 24043884 Free PMC article.
-
Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies.J Virol. 2015 Apr;89(8):4227-36. doi: 10.1128/JVI.00154-15. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631085 Free PMC article.
-
Dengue Virus Control of Type I IFN Responses: A History of Manipulation and Control.J Interferon Cytokine Res. 2015 Jun;35(6):421-30. doi: 10.1089/jir.2014.0129. Epub 2015 Jan 28. J Interferon Cytokine Res. 2015. PMID: 25629430 Free PMC article. Review.
-
Different STAT Transcription Complexes Drive Early and Delayed Responses to Type I IFNs.J Immunol. 2015 Jul 1;195(1):210-216. doi: 10.4049/jimmunol.1401139. Epub 2015 May 27. J Immunol. 2015. PMID: 26019270 Free PMC article.
-
How Dengue Virus Circumvents Innate Immunity.Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review.
Cited by
-
Development of a transmission model for dengue virus.Virol J. 2013 Apr 23;10:127. doi: 10.1186/1743-422X-10-127. Virol J. 2013. PMID: 23617898 Free PMC article.
-
Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes.Curr Opin Immunol. 2012 Aug;24(4):364-78. doi: 10.1016/j.coi.2012.04.011. Epub 2012 May 30. Curr Opin Immunol. 2012. PMID: 22651901 Free PMC article. Review.
-
Basal interferon signaling and therapeutic use of interferons in controlling rotavirus infection in human intestinal cells and organoids.Sci Rep. 2018 May 29;8(1):8341. doi: 10.1038/s41598-018-26784-9. Sci Rep. 2018. PMID: 29844362 Free PMC article.
-
Animal models for dengue vaccine development and testing.Clin Exp Vaccine Res. 2017 Jul;6(2):104-110. doi: 10.7774/cevr.2017.6.2.104. Epub 2017 Jul 26. Clin Exp Vaccine Res. 2017. PMID: 28775974 Free PMC article. Review.
-
The molecular basis for differential type I interferon signaling.J Biol Chem. 2017 May 5;292(18):7285-7294. doi: 10.1074/jbc.R116.774562. Epub 2017 Mar 13. J Biol Chem. 2017. PMID: 28289098 Free PMC article. Review.
References
-
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. - PubMed
-
- van den Broek MF, Muller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;148:5–18. - PubMed
-
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. - PubMed
-
- Schindler C, Strehlow I. Cytokines and STAT signaling. Adv Pharmacol. 2000;47:113–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

