HDACs link the DNA damage response, processing of double-strand breaks and autophagy
- PMID: 21368826
- PMCID: PMC3935290
- DOI: 10.1038/nature09803
HDACs link the DNA damage response, processing of double-strand breaks and autophagy
Abstract
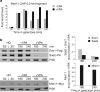
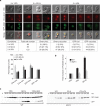
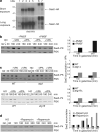
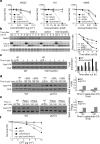
Protein acetylation is mediated by histone acetyltransferases (HATs) and deacetylases (HDACs), which influence chromatin dynamics, protein turnover and the DNA damage response. ATM and ATR mediate DNA damage checkpoints by sensing double-strand breaks and single-strand-DNA-RFA nucleofilaments, respectively. However, it is unclear how acetylation modulates the DNA damage response. Here we show that HDAC inhibition/ablation specifically counteracts yeast Mec1 (orthologue of human ATR) activation, double-strand-break processing and single-strand-DNA-RFA nucleofilament formation. Moreover, the recombination protein Sae2 (human CtIP) is acetylated and degraded after HDAC inhibition. Two HDACs, Hda1 and Rpd3, and one HAT, Gcn5, have key roles in these processes. We also find that HDAC inhibition triggers Sae2 degradation by promoting autophagy that affects the DNA damage sensitivity of hda1 and rpd3 mutants. Rapamycin, which stimulates autophagy by inhibiting Tor, also causes Sae2 degradation. We propose that Rpd3, Hda1 and Gcn5 control chromosome stability by coordinating the ATR checkpoint and double-strand-break processing with autophagy.
Figures

Comment in
-
Molecular biology: The expanding arena of DNA repair.Nature. 2011 Mar 3;471(7336):48-9. doi: 10.1038/471048a. Nature. 2011. PMID: 21368822 No abstract available.
Similar articles
-
Molecular biology: The expanding arena of DNA repair.Nature. 2011 Mar 3;471(7336):48-9. doi: 10.1038/471048a. Nature. 2011. PMID: 21368822 No abstract available.
-
Sae2 antagonizes Rad9 accumulation at DNA double-strand breaks to attenuate checkpoint signaling and facilitate end resection.Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):E11961-E11969. doi: 10.1073/pnas.1816539115. Epub 2018 Dec 3. Proc Natl Acad Sci U S A. 2018. PMID: 30510002 Free PMC article.
-
Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2.Nucleic Acids Res. 2015 Aug 18;43(14):6889-901. doi: 10.1093/nar/gkv520. Epub 2015 May 27. Nucleic Acids Res. 2015. PMID: 26019182 Free PMC article.
-
Acetylation: a novel link between double-strand break repair and autophagy.Cancer Res. 2012 Mar 15;72(6):1332-5. doi: 10.1158/0008-5472.CAN-11-3172. Cancer Res. 2012. PMID: 22422989 Review.
-
Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks.DNA Repair (Amst). 2013 Oct;12(10):791-9. doi: 10.1016/j.dnarep.2013.07.009. Epub 2013 Aug 13. DNA Repair (Amst). 2013. PMID: 23953933 Review.
Cited by
-
Implications of endoplasmic reticulum stress and autophagy in aging and cardiovascular diseases.Front Pharmacol. 2024 Jul 25;15:1413853. doi: 10.3389/fphar.2024.1413853. eCollection 2024. Front Pharmacol. 2024. PMID: 39119608 Free PMC article. Review.
-
The emerging role of acetylation in the regulation of autophagy.Autophagy. 2013 Jun 1;9(6):819-29. doi: 10.4161/auto.23908. Epub 2013 Mar 6. Autophagy. 2013. PMID: 23466676 Free PMC article. Review.
-
USP14 is a deubiquitinase for Ku70 and critical determinant of non-homologous end joining repair in autophagy and PTEN-deficient cells.Nucleic Acids Res. 2020 Jan 24;48(2):736-747. doi: 10.1093/nar/gkz1103. Nucleic Acids Res. 2020. PMID: 31740976 Free PMC article.
-
Base excision repair AP endonucleases and mismatch repair act together to induce checkpoint-mediated autophagy.Nat Commun. 2013;4:2674. doi: 10.1038/ncomms3674. Nat Commun. 2013. PMID: 24154628 Free PMC article.
-
Sulforaphane Decrease of SERTAD1 Expression Triggers G1/S Arrest in Breast Cancer Cells.J Med Food. 2019 May;22(5):444-450. doi: 10.1089/jmf.2018.4195. J Med Food. 2019. PMID: 31084542 Free PMC article.
References
-
- Bird AW, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. - PubMed
-
- Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. - PubMed
-
- Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

